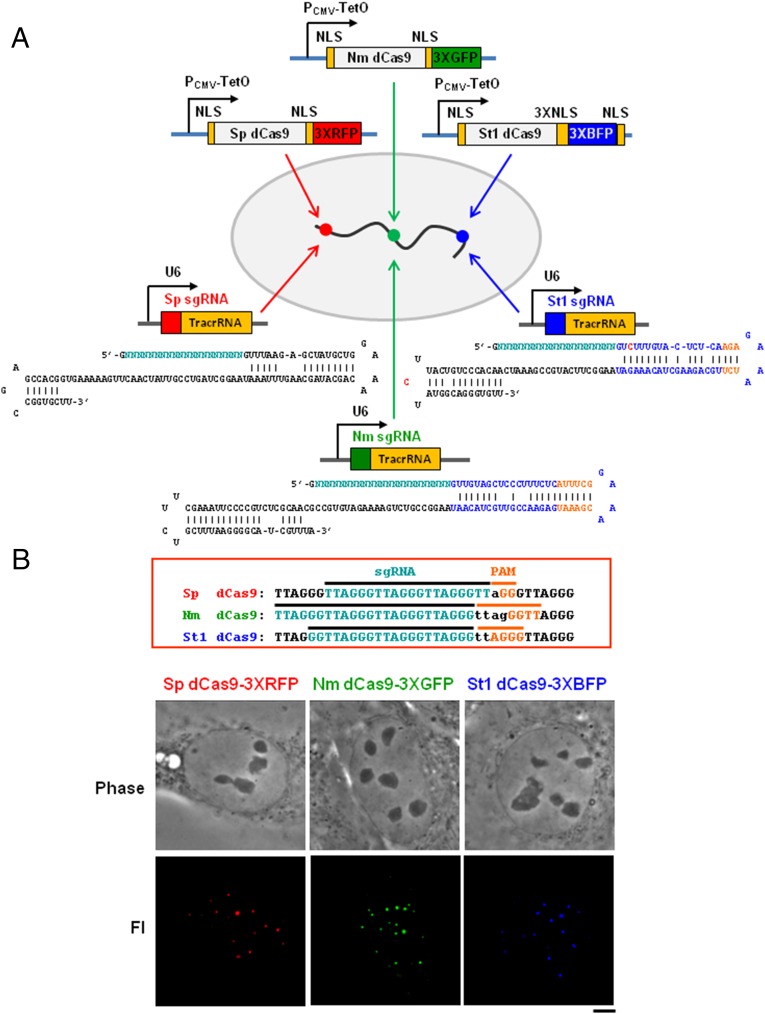
Fig. 1.
The multicolor CRISPR labeling system exploiting Cas9 orthologs. (A) Designs of dCas9 (“d” for “nuclease-dead”) ortholog-fluorescent proteins: S. pyogenes Sp dCas9, N. meningitidis Nm dCas9 and S. thermophilus St1 dCas9 fused to NLSs and under the control of the CMV-TetO promoter. The cognate sgRNAs Sp sgRNA, Nm sgRNA, and St1 sgRNA are under the control of the U6 promoter, and their sequences are shown below, with the mutations in red and hairpin extensions in orange. Directed by the appropriate sgRNA, different chromosomal loci are expected to become fluorescently labeled with any one of the three spectral versions of dCas9. In the diagram, such hypothetical targeted sequences are indicated by the color matching a particular dCas9/sgRNA ortholog. (B) sgRNAs and PAM sequences designed for targeting telomeres by each of the dCas9 orthologs Sp, Nm, and St1. The panels below are phase-contrast (Upper) and fluorescent signals for telomeres (Lower) images for human U2OS cells expressing the indicated dCas9-FPs and cognate sgRNAs. (Scale bar: 5 μm.)