Figure 1.
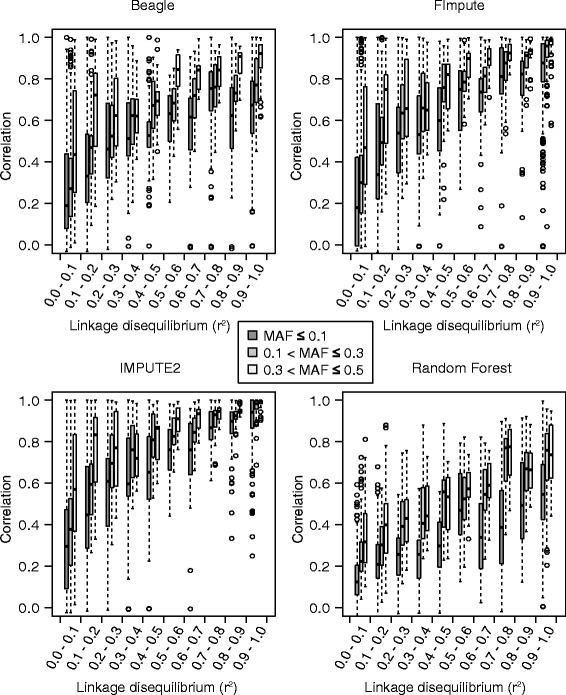
Linkage disequilibrium influences the accuracy of imputing missing values. The relationship between linkage disequilibrium (as measured by r2 between 90 k SNPs and the respective most closely linked 9 k SNPs) and the average correlation between observed and imputed genotypic data, as calculated using map-dependent (Beagle, FImpute, and IMPUTE2) and map-independent (Random Forest) imputation algorithms, for a reference population size of 50 out of 371 lines. Trends are shown as boxplot displays separately for three minor allele frequency (MAF) classes.
