Figure 3.
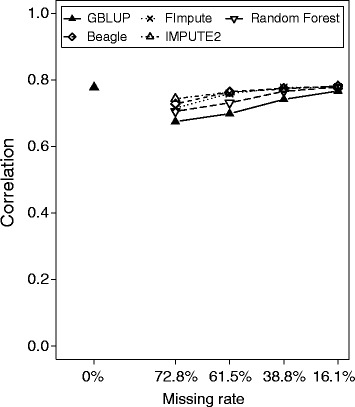
Imputing improves accuracy of prediction of genomic selection based on GBS-like data sets. Correlation between true and predicted genotypic values applying genomic selection for the original 90 k SNP data set (GBLUP, 0%), as well as for genotype-by-sequencing-like data sets where missing values were not imputed (GBLUP) or were imputed with map-dependent (Beagle, FImpute and IMPUTE2) and map-independent (Random Forest) algorithms for rates of missing values of 72.8%, 61.5%, 38.8% and 16.1% in the total population of 371 lines.
