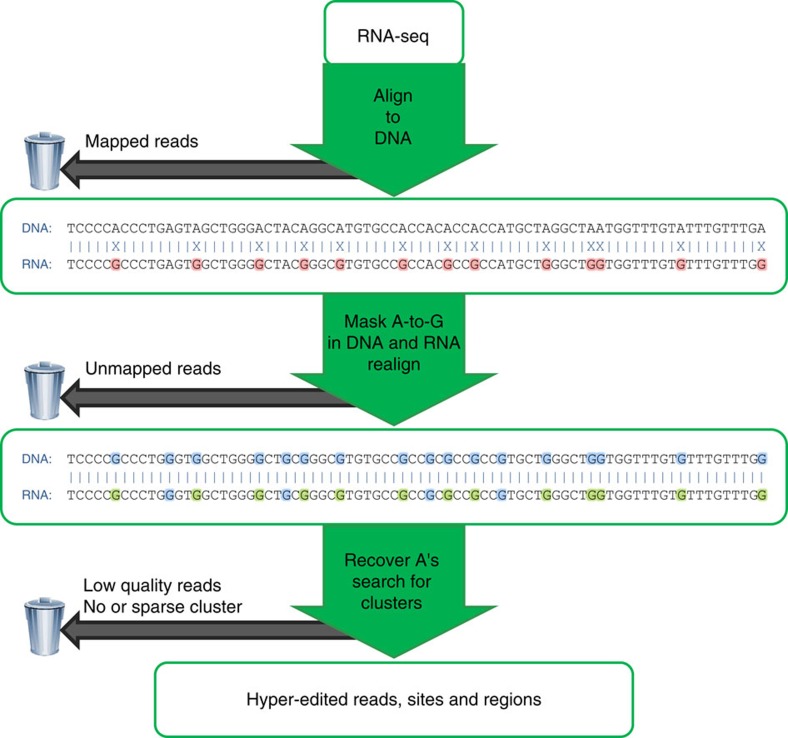
Figure 1. Overview of our hyper-editing detection pipeline.
In the first step, an RNA-seq data set is aligned to the reference genome and mapped reads are discarded. Hyper-edited reads do not map in this step since they contain too many mismatches (see a hypothetical example in the top alignment; edited nucleotides highlighted in red). To detect potential RNA editing in the unmapped reads, we realign them after masking the editing sites by transforming all As to Gs in both RNA and DNA (bottom alignment; transformed nucleotides in blue, edited nucleotides in green). For those reads that realign, the original sequences are recovered and examined for high-quality clusters of A-to-G mismatches. Reads with large, dense clusters of A-to-G differences are designated as hyper-edited, and their editing sites are recorded. See complete details in Methods.