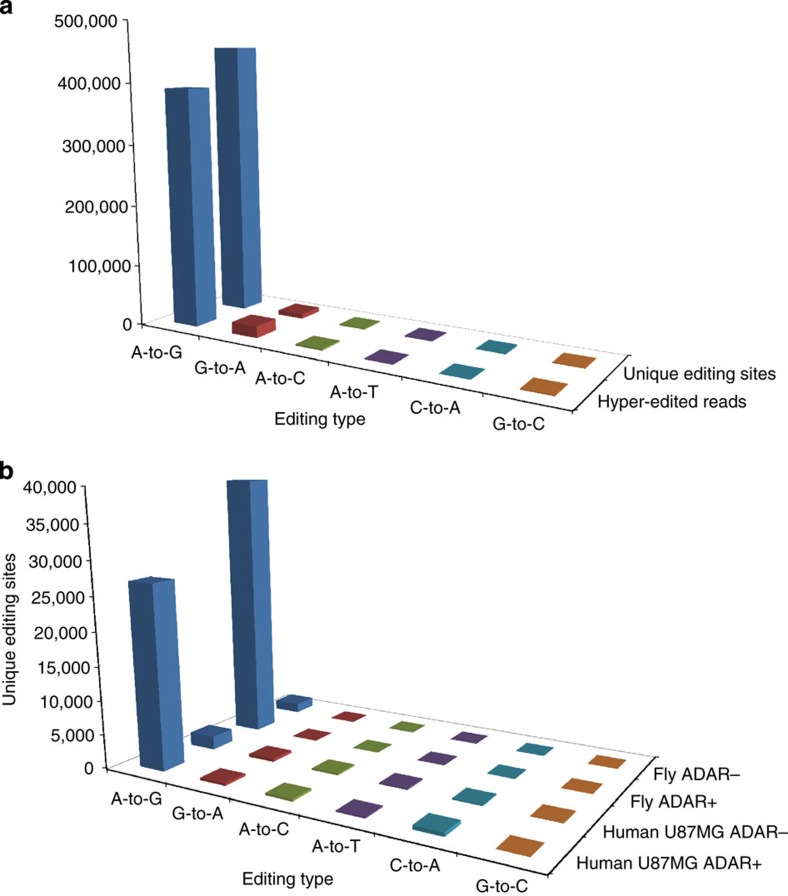
Figure 2. Counts of hyper-editing events.
(a) Hyper-editing identified in the Illumina BodyMap 75-bp SE read set. Most of the detected hyper-edited reads (390,881/414,898; 94.2%) were of A-to-G type. A total of 455,014 unique A-to-G-editing sites were discovered, 97.2% of all the detected unique sites. (b) Most editing sites were eliminated in ADAR− samples. We detected hyper-editing sites in Drosophila nascent-RNA-Seq data from either wild-type (ADAR+) flies or from ADAR-null (ADAR−) flies. The number of A-to-G sites detected in the ADAR+ sample was ≈20-fold larger than in the ADAR− sample (39,472 versus 1,436; there were 2 × 10−4 editing sites per (mapped) read in ADAR+ compared with 9 × 10−6 in ADAR−). Similarly, we detected hyper-editing sites in the human U87MG cell line, either with (ADAR−) or without (ADAR+) siRNA-induced silencing of ADAR1. The number of sites in the ADAR+ sample was much higher than in the ADAR− sample (27,124 versus 1,992, or 3 × 10−4 versus 2 × 10−5 sites per mapped read). The number of non-A-to-G-editing sites is also presented. In the ADAR+ samples, more than 94% of the detected sites were A-to-G. In the ADAR− samples, while the number of A-to-G sites significantly decreased, the counts of the other mismatches were almost indifferent to the absence of ADARs.