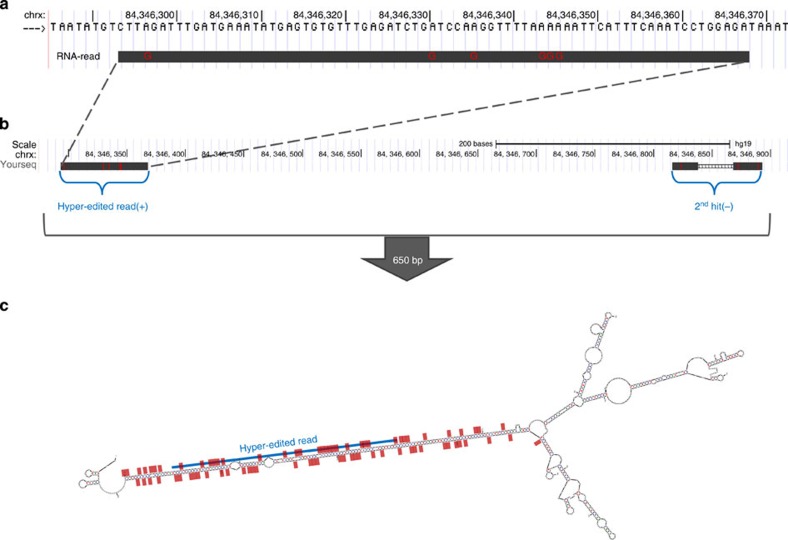
Figure 4. Hyper-editing in a region outside known repeats.
(a) An alignment of a hyper-edited read to a DNA region outside known repeats. (b) A BLAT search66 (The UCSC Genome Browser62), using the hyper-edited read as a query, returned two hits in an opposite orientation: one at the hyper-edited region and another ≈650 bp away, allowing the potential formation of dsRNA. (c) Prediction of the secondary structure of the entire 650-bp region (mfold67). The blue line shows the location of the hyper-edited read of a; red dots are located at editing sites detected along the entire 650-bp region (both from the read shown at a as well as from other reads). All editing sites are lined along a dsRNA structure. Experimental validation of many of those predicted editing sites is described in Methods and in Supplementary Fig. 3a.