Abstract
Recent changes in the Fungal Code of Nomenclature and developments in molecular phylogeny are about to lead to dramatic changes in the naming of medically important molds and yeasts. In this article, we present a widely supported and simple proposal to prevent unnecessary nomenclatural instability.
ONE FUNGUS, ONE NAME
Until recently, polymorphic higher fungi (Dikarya) were allowed to carry multiple names describing sexual (teleomorph) and various asexual (anamorph) stages of their life cycles. These stages could develop independently from each other, and their genetic relationship was often difficult to establish. Today, with the wide application of molecular methods, this problem has largely been solved. With the introduction of molecular genetic approaches, the dual naming system is no longer necessary. Two international expert symposia recently held in Amsterdam, The Netherlands, have been devoted to the fate of dual naming in fungi: One Fungus = One Name symposium on 19 and 20 April 2011 and One Fungus = Which Name symposium on 12 and 13 April 2012. The resulting Amsterdam Declaration on Fungal Nomenclature (1) requested the abolition of Article 59 of the Code of Botanical Nomenclature, the provision that sanctioned multiple names for the same fungus. Under the new Code of Nomenclature of Algae, Fungi and Plants, from 1 January 2013, this system is no longer permitted. One of the consequences of the declaration is that the criteria for naming fungi have changed entirely.
The dual naming system had been useful during the age of the microscope. Today, the main criteria for classification have moved from phenotype to genotype. Analysis of nucleic acid sequence variation now guides taxonomy and has replaced phenotype with the history of phylogenetic relationships and, occasionally, sexual compatibility. The first phase began after the introduction of PCR in the late 1980s and resulted in the discovery of new species by concordance of gene genealogies in several pathogenic fungi. This is now being expanded following the advent of next-generation sequencing, which is discovering genetically distinct populations that deserve species status. The newly discovered species are genealogically distinct but cryptic in the sense that they were not suspected from morphological phenotype—although after they are recognized, distinguishing phenotypes may be discovered later on. In addition, taxonomy of environmental fungi is developing at a very rapid pace, which has a profound impact on nomenclature of opportunistic fungi. Anatomic morphological categories, such as coelomycetes or hyphomycetes, have become redundant, which implies that all mycological textbooks have become obsolete. Diagnostic laboratories will have to change the type and interpretation of data used to identify fungi. The changes will hopefully contribute to nomenclatural stability in the future, but it is obvious that this will not be achieved before the end of a transition phase. Although the shift in identification can be viewed as simply a change in technique from microscopy to DNA sequence analysis, it must also be viewed as a major intellectual shift to a system based on evolution as inferred from comparison of genotypes.
Coincidently with this process, a significant expansion of the number of etiologic agents of disease is noticed. In the literature, it is often stated that (i) this growth is especially due to the increasing population of immunocompromised patients. A more pertinent cause, however, is (ii) the development of our knowledge, driven by easy access to sequencing technology. The advent of population genomics, which increases the sampled diversity by as many as 4 orders of magnitude, can only increase this trend. A further source of new clinical species to be recognized in diagnostic laboratories is the fact that (iii) also sterile or nonculturable fungi can now be classified according to sequence analysis of PCR-generated amplicons. The expansion of the number of clinically relevant fungi is clearly demonstrated in the Atlas of Clinical Fungi of which the first edition (2) contained 320 species, while the 2013 edition of the same book (3) counts a staggering 560 species, and the number of species is still growing at the same pace.
When only a single name should be used for these fungi, most of which bear several names at present, the question is which name has priority? One of the principles of nomenclature is to choose (i) the oldest name, either anamorph or teleomorph, which is also mostly the most widely applied name. The new code has the second rule that if both anamorph and teleomorph names have been widely used, (ii) the teleomorph name is to be maintained unless a formal application in favor of the anamorph name has been made. Both choices are dependent on (iii) the size of the genus and its rank in the taxonomic hierarchy.
CHANGES AT THE GENUS LEVEL AND ABOVE
Starting with item iii, genus concepts are related to the amount and diversity of available biological material. Nucleic acid variation has provided a means of ensuring that genera are monophyletic, which is a leading principle of modern taxonomy. Species differing at the ordinal or even family level are no longer accepted as members of a single genus. Hence, orders and sometimes families are taxonomically relevant entities. The advantage of the phylogenetic approach is that close relatives come together even if they are morphologically quite different; this may be useful for predictions of pathogenicity or antifungal susceptibility. Conversely, distant relationships are expected to predict large differences in clinically relevant parameters. Until today, most clinical fungi had been grouped in anamorphic form genera based on phenotype that do not necessarily represent phylogenetic relatedness. For example, non-albicans Candida species comprise a random mixture of species that have been grouped together only because they are morphologically indistinguishable and physiologically similar, but some of them are as distant from each other as humans are from frogs. Applying names that acknowledge this diversity would be logical and medically meaningful. Candida albicans and Candida glabrata are evolutionary distant species with different antifungal susceptibilities (4). Penicillium marneffei is unrelated to most of the saprobic Penicillium species, but it belongs to a group of penicillium-like species classified in the genus Talaromyces that possess similar virulence factors. Therefore, reclassification of P. marneffei in Talaromyces (5) is useful for the medical mycologist. Similar reasoning can be applied to categories higher up in the fungal system. Although the Zygomycota appears not to be monophyletic, there has been reluctance to abandon the name, but as the diseases caused by the main groups composing it, the Mucoromycotina and Entomophthoromycotina, are fundamentally different (6), their separation would be a step forward. The novel approach using phylogeny as a main criterion enhances information content of the taxonomy hierarchy.
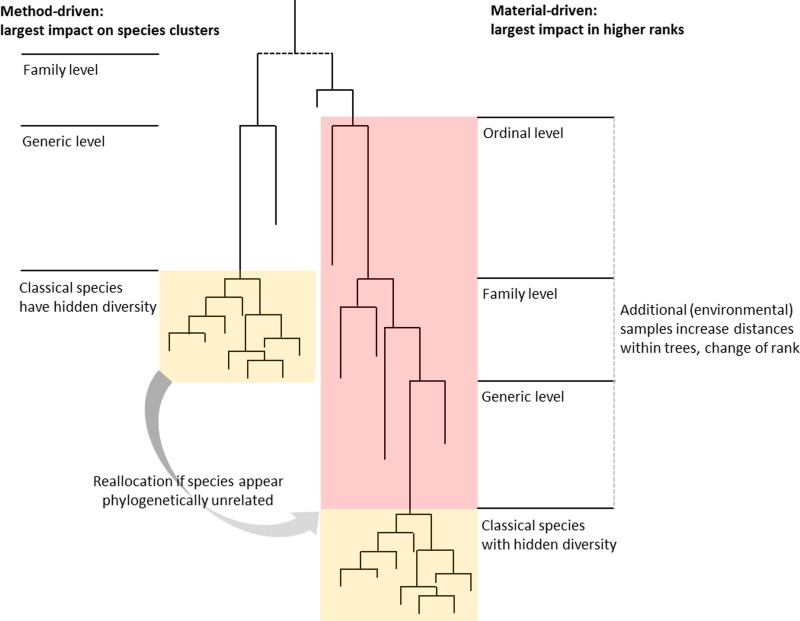
The above is valid on the assumption that the “real” phylogenetic tree of the fungal kingdom is known. However, it should be realized that only a minor fraction of the existing fungal diversity has been described thus far. Large numbers of novel species are continuously being discovered due to the exploration of new habitats. The main cause of generic instability is material driven; phylogenetic trees are highly sensitive to taxon sampling effects (Fig. 1), and this situation will remain for many decades to come. Description of species on the basis of single sequences from metagenomic data will further complicate the taxonomic system (7). The basis of a genus or of any higher rank in the taxonomic hierarchy is a monophyletic branch of an underlying phylogenetic tree, replacing phenotypic techniques. Fungi may appear to belong to other phylogenetic groups than hypothesized earlier.
FIG 1.
Diagram of name changes driven by methodical advances and sampling effects: subdivisions, reallocations, and rank inflations.
The clade approach for naming species, genera, and above has fundamental shortcomings, because of the comparative nature of data and also because no delimitation criterion exists. Determination of all higher taxonomic ranks when they are defined exclusively by sequence data is inherently arbitrary and therefore unstable, leading to numerous transfers of species from one genus to the next. Phylogeny deepens our understanding of the fungal kingdom, but phylogenetic trees are just an approximation of the truth. During the last 10 years, more name changes have been proposed for medical fungi than in the previous 70 years; examples can be found in Table 1. Many clades in a tree can be statistically supported, but what is the level of diversity to recognize a genus? At present, Aspergillus contains about 290 species, while genera in the Scedosporium lineage, with comparable levels of bar coding gaps between neighboring taxa, contain only 1 to 6 species. If genera become nearly congruent to species, then the genus becomes a redundant category.
TABLE 1.
Some examples of medically important fungi that have undergone recent multiple changes, with year of publication and reasons for rearrangement
| Original name in medicine | Transitional name(s) | Current name |
|---|---|---|
| Hendersonula toruloidea 1933 | Scytalidium hyalinum 1977 (supposed synonymy) | Neoscytalidium dimidiatum 2006 (phylogenetic rearrangement) |
| Scytalidium dimidiatum 1989 (earlier synonym Torula dimidiata 1887) | ||
| Nattrassia mangiferae 1989 (supposed identity of coelomycete anamorph) | ||
| Fusicoccum dimidiatum (2005) (phylogenetic rearrangement) | ||
| Neofusicoccum mangiferae 2006 (phylogenetic rearrangement) | ||
| Neoscytalidium hyalinum 2013 (phylogenetic rearrangement) | ||
| Trichosporon capitatum 1942 | Geotrichum capitatum 1977 (phylogenetic rearrangement) | Saprochaete capitata 2004 (phylogenetic rearrangement of anamorph) |
| Blastoschizomyces capitatus 1985 (synanamorph genus) | Magnusiomyces capitatus 2004 (phylogenetic rearrangement of teleomorph; priority of either genus name still to be established) | |
| Blastoschizomyces pseudotrichosporon 1982 (synonymy) | ||
| Dipodascus capitatum 1996 (description of teleomorph) | ||
| Candida utilis 1952 | Hansenula jadinii 1979 (conspecificity with Candida utilis, description of teleomorph) | Cyberlindnera jadinii 2009 (nomenclatural correction) |
| Pichia jadinii 1984 (phylogenetic rearrangement) | ||
| Lindnera jadinii 2008 (phylogenetic rearrangement) | ||
| Allescheria boydii 1922 | Pseudallescheria boydii (teleomorph) | Scedosporium boydii (consensus chosen by Scedosporium community, but Pseudallescheria teleomorph genus name has been prioritized by nomenclatural community) |
| Cephalosporium boydii 1922 | Scedosporium boydii (prevalent anamorph; name for third morph neglected) | |
| Dendrostilbella boydii 1922 (anatomic names for a trimorphic fungus) |
A profoundly debated example is the anamorph genus Aspergillus. The Aspergillus genus was discovered by Micheli in 1753, based on Aspergillus glaucus. However, the medically important fungus Aspergillus fumigatus is a member of another phylogenetic clade in Aspergillus that also bears the teleomorph name Neosartorya. In the case of A. fumigatus, which was found to have a Neosartorya sexual state in 2009 after a concerted effort to generate ascocarps (8), the anamorph name A. fumigatus is older and more widely used than N. fumigata. However, the A. fumigatus/N. fumigata clade is phylogenetically remote and phenotypically distinct from the clade that contains the generic type, A. glaucus (9). There are two proposals in the literature. One advocates making Aspergillus a very large genus covering numerous clades, including those of A. glaucus and A. fumigatus (9). After careful discussion among the members, this proposal was chosen by the International Commission of Penicillium and Aspergillus. The other advocates applying existing teleomorph names to all monophyletic branches except one, and that clade alone would retain the name Aspergillus (10).
Both proposals have their pros and cons. In the first case, Aspergillus would remain the name for all, but as the broad phylogeny includes genera such as Phialosimplex and Polypaecilium would have to be renamed in Aspergillus. The breadth of phenotype embraced by Aspergillus would include fungi that have never been associated with Aspergillus, but the phylogenetic data supporting this are solid (11), and it should be acknowledged that aspergilli with deviating morphological features do exist. In the second case, A. fumigatus would be named Neosartorya fumigata because Neosartorya species form a distinct monophyletic clade within the exiting genus Aspergillus. This may be unpleasant for medical mycologists, but for many other users of the fungal kingdom, this is good news as Aspergillus would then be used to refer specifically to Aspergillus species in the subgenus Circumdati. These include fungi with even more prominent economic value, for example the Asian food fungus, Aspergillus oryzae, its close relative, the aflatoxin producer, Aspergillus flavus, and the industrial fermentation workhorse, Aspergillus niger.
In summary, a major source of potential name changes, in addition to problems of dual nomenclature, is linked to phylogeny as a leading principle, while phylogenetic trees and taxonomic hierarchies are still under construction and subject to change while nature's diversity is better understood.
CHANGES AT THE SPECIES LEVEL AND BELOW
Species recognition is primarily method driven, i.e., tied to the method of observation. The shift from recognizing species by observable phenotype to recognizing them by nucleic acid variation has resulted in a proliferation in the number of species. In recent years, more precision has been achieved in molecular methods by replacing species recognition in single-gene studies by multiple-gene analysis of lineages and populations, leading to molecularly defined taxa (sibling or cryptic species; Table 2). This trend will continue now that whole genomes are becoming available for large numbers of strains within a single species, as has been shown with model fungi (12–14).
TABLE 2.
Some taxonomic definitions appropriate for medical mycology that are used in the present paper
| Term | Taxonomic definition |
|---|---|
| Species complex | A monophyletic clade of species with equivalent clinical relevance |
| Sibling species | Species that share the same, most recent common ancestor |
| Cryptic species | Species recognized by nucleic acid variation that had not been recognized as distinct by morphological phenotypes. Once recognized, phenotypic characters useful for identification may be discovered in the future. |
| (Sub)clade/monophyletic group | Phylogenetic group consisting of an ancestral species and all its descendants. Clades and subclades can be recognized at any given taxonomic level. Statistical tests are used to gauge the support for these groups. |
| Lineage | Series of species connected by evolutionary descent, not necessarily representing all known descendants |
| Cluster/group | Terminal series of phylogenetically related species, used when precise relationships are uncertain. |
| Type | Entity defining a taxonomic name and indicated as such in the protologue. Species and below are defined by a specimen, whereas higher taxonomic entities are defined by the first lower category. |
| Neotype | New specimen in accordance with the protologue in case the original type material is lost. |
| Epitype | Reference specimen accordance with the protologue when the original material is not interpretable. |
| Protologue | Original description and any other representation of a taxonomic entity. |
When this process is clinically relevant, the novel naming system should rapidly be adopted. For example, Sporothrix schenckii, agent of human sporotrichosis, contains a hypervirulent sibling now known as Sporothrix brasiliensis causing large epidemics in Brazil (15). In another example, Scedosporium aurantiacum, one of the novel species recognized within the former umbrella species Scedosporium apiospermum is significantly less susceptible to currently used antifungal agents than the original species (16).
Application of more-variable genes to the same fungi will always lead to discovery of more diversity, again as demonstrated by population genomics. Almost all fungal species that have been examined show evidence of recombination and, therefore, exhibit upper and lower bounds to species recognition. The lower bound has been found using population genomic analysis to identify genetically differentiated populations of interbreeding individuals, and development of the upper bound has been observed where species have evolved reinforced barriers to mating in some areas of sympatry and not others (17). Only in clonal species would these bounds not apply and entities can be subdivided ad infinitum. In practice, many, perhaps most, fungal lineages combine enough recombination with clonal behavior to be constrained by both upper and lower species boundaries. There is no gold standard as a measure of taxonomic diversity at the species level, and thus optimal barcoding genes are differentially effective between different groups.
Differences between molecular siblings may not always have clinical relevance. Distinction of cryptic species may then be taxonomically valid and scientifically meaningful but remain undetectable in routine laboratory analyses. On a global scale of daily clinical diagnostics, attempting to detect these species would require an investment that would not contribute to patient care and is therefore not recommended until further research provides justification of these additional efforts. For example, Aspergillus niger was recently found (18) to contain a molecular sibling, Aspergillus awamori. Varga et al. (19) explicitly mention that there are no differences in phenotypic characteristics, such as metabolite profiles between the two, and clinical differences between the two species have yet to be discovered. As difficult as it might be to distinguish two species, imagine the case of the ubiquitous contaminant Cladosporium cladosporioides, which today contains 39 cryptic species (20), 38 of which have never been proven as agents of human infection.
Neighboring siblings that are identical in patient management and normally characterized phenotype might in routine diagnostics better be taken together as “complexes.” A “species complex” of medically important fungi would be considered a cluster of cryptic species that are clinically identical. This is the current indication of series of closely related Fusarium species (21), and similar approaches have been adopted for Cladosporium (20) and elsewhere. The word “complex” has no nomenclatural status and does not require any name change. Species complexes can nevertheless be sharply delimited and validated by molecular data. Diagnostic markers can be developed for the molecular siblings and for the species complex. If an author wishes to describe other, less clearly defined species diversities, terms like “group,” “cluster,” “lineage,” and “clade” are available (Table 2).
In summary, a major source of name changes at the species level is increased precision of molecular techniques; this nomenclatural instability is thus largely method driven. The distinguished entities, even when scientifically correct and meaningful, may not always have clinical relevance, although this significance may perhaps be discovered in the future.
CONCLUSIONS AND PROPOSALS
During the process of naming each fungus, teleomorph and anamorph names as well as their synonyms are being considered in a way that would increase acceptance and stability. Even strict nomenclatural rules provide a certain degree of liberty. Many of the classical medical fungi and many of their synonyms were described a long time ago, and type material is often lacking or uninterpretable. The present paper does not argue for one solution or the other in any of the examples above but simply notes that there is more than one way to apply one name for one fungus. Nomenclatural changes of medically important fungi usually take decades to gain wide acceptance. Any change may be viewed as a process taking place in the community, rather than as a singular result of a phylogenetic study. Taxonomy is a dynamic science, which cannot be muzzled by nomenclatural protocols. Reallocations, rank changes, and generic disarticulations or reunifications will remain common practice. Theoretically, the taxonomic system should reflect the true phylogeny of the fungal kingdom, but obviously we have not yet reached that stage. Additional data and techniques improving the system are continually generated and published. Therefore, it may be more prudent to wait until a larger degree of stability and consensus is achieved. Many names of opportunistic fungi are published as part of studies on environmental fungi, where genera might comprise dozens or hundreds of species. The genus name is linked to its type species, and if this species appears to be different from all the others, all remaining names need to be recategorized in another genus. The result is that the number of name changes can be tremendous—a compelling reason to be careful with reallocations where scientific support is still fragmentary.
How should the field of medical mycology treat the new diversity seen in the taxonomy of medically important fungi? At the genus level, these are reallocations, rank changes, and generic disarticulations or reunifications that stem from studies of more materials and from having to choose one name from two or more current names. Where examination of new materials suggests new names, we urge taxonomists to delay introduction of new names until they have sampled sufficient material. Where name change results from having to choose one of several names, maintaining taxa that have similar medical attributes would serve medical mycology; such taxa should neither be so big as to hide medically important phenotypic variation nor so small to lessen the distinction between genera and species. Where the names of medically important fungi do change—and many will change as the new code is applied—it may reflect the fact that medical mycology is just one of many socially important activities that focus on fungi. Good taxonomic studies do not always need new names immediately. For clinical routine, it is advocated to follow changes with some delay, after validation by a convincing body of data, until a sufficient degree of stability and consensus is reached.
At the species level, most changes concern subdivisions of classical phenotypic species, as new methods allow mycologists to examine more and more of the genetic variation, including the entire genomes of populations of fungi. As new research techniques become widespread in clinics, clinicians also will be able to recognize more species. Until, after clinical evaluation, newly discovered species prove to be different in terms of patient management, in daily clinical routine, it might be better to unite such siblings as “species complexes.”
Researchers and clinicians should work together to achieve a reasonable degree of nomenclatural stability during the decades when large changes are unavoidable. Even when a disease caused by two or more closely related species is treated by the same therapy, insights into diversity of the species may advance medicine by allowing clinicians to use this knowledge to discover previously overlooked and consistent differences. Specialized websites are available where diagnostic materials are provided as an aid for identification and the best current taxonomy is available as matters change.
ACKNOWLEDGMENTS
The contents of this paper have been communicated to the Nomenclature Committee for Fungi (NCF) (www.ima-mycology.org/CFF): S. A. Redhead, L. Norvell, and P. Kirk, and the International Commission on the Taxonomy of Fungi (ICTF): K. A. Seifert and A. Miller.
The members of the ISHAM Working Group on Nomenclature of Medical Fungi are Teun Boekhout (CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands, and Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, The Netherlands), Arunaloke Chakrabarti (Mycology Division, Department of Medical Microbiology, Postgraduate Institute of Medical Education and Research [PGIMER], Chandigarh, India), Anuradha Chowdhary (Vallabhbhai Patel Chest Institute, Delhi, India), Garry Cole (Department of Biology and South Texas Center for Emerging Infectious Diseases, University of Texas at San Antonio, San Antonio, TX), Olivier A. Cornely (Department I of Internal Medicine, University Hospital, ZKS Köln, BMBF 01KN1106, Cologne and Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases [CECAD], University of Cologne, Cologne, Germany), Pedro W. Crous (CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands, and Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, The Netherlands), Christophe D’Enfert (Fungal Biology and Pathogenicity Unit, Institut Pasteur, Paris, France), Dea Garcia-Hermoso (Institut Pasteur, Unité de Mycologie Moléculaire, Centre National de Référence Mycoses Invasives et Antifongiques, Paris, France), D. David Ellis (School of Molecular & Biomedical Science, The University of Adelaide, Adelaide, Australia, and National Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD), Cornelia Lass-Flörl (Division of Hygiene and Medical Microbiology, Innsbruck Medical University, Innsbruck, Austria), Stuart Levitz (Center of AIDS Research, University of Massachusetts Medical School, Worcester, MA), Ruo-Yu Li (Research Center for Medical Mycology, Peking University, Beijing, China), Aaron P. Mitchell (200B Mellon Institute, Department of Biological Sciences, Carnegie Mellon University, Pittsburgh, PA), Kerry O’Donnell (Bacterial Foodborne Pathogens & Mycology Research Unit, U.S. Department of Agriculture, Agriculture Research Service, Peoria, IL), John R. Perfect (Department of Medicine, Division of Infectious Diseases, Duke University Mycology Research Unit [DUMRU], Durham, NC), Flavio Queiroz Telles (Division of Infectious Diseases, Department of Public Health, Hospital de Clinicas, Universidade Federal do Parana, Curitiba, Brazil), Deanna A. Sutton (Fungus Testing Laboratory, Department of Pathology, School of Medicine, University of Texas Health Science Center at San Antonio, San Antonio, TX), Kerstin Voigt (Microbial Resource Collection Friedrich-Schiller-Universität, Jena, Germany), Theodore C. White (School of Biological Sciences, University of Missouri at Kansas City, Kansas City, MO), and Liyan Xi (Department of Dermatology, Second Affiliated Hospital, Sun Yat-Sen University, Guangzhou, China).
Biography

Sybren de Hoog (born 1948) is group leader of Medical Fungi at the CBS-KNAW Fungal Biodiversity Centre at Utrecht, The Netherlands. He also holds professorships at universities in The Netherlands, Brazil, China, and Saudi Arabia. Areas of interest are ecology and evolution of pathogenic fungi, on which he has written over 550 refereed papers. He is the leading author of the Atlas of Clinical Fungi. Dr. de Hoog is past-President of the International Society for Human and Animal Mycology (ISHAM). In this function, he assisted in the organization of ISHAM and satellite congress in Tokyo, Japan, and Beijing, China, in 2009, and he was program chairman of the TIMM congress in Amsterdam, The Netherlands. His teaching activities include the CBS Course Medical Mycology for hospital personnel, which is given regularly in several European and Asian countries.
Footnotes
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, Taylor JW, Wingfield MJ, Abaci O, Aime C, Asan A, Bai FY, de Beer ZW, Begerow D, Berikten D, Boekhout T, Buchanan PK, Burgess T, Buzina W, Cai L, Cannon PF, Crane JL, Damm U, Daniel HM, van Diepeningen AD, Druzhinina I, Dyer PS, Eberhardt U, Fell JW, Frisvad JC, Geiser DM, Geml J, Glienke C, Gräfenhan T, Groenewald JZ, Groenewald M, de Gruyter J, Guého-Kellermann E, Guo LD, Hibbett DS, Hong SB, de Hoog GS, Houbraken J, Huhndorf SM, Hyde KD, Ismail A, Johnston PR, Kadaifciler DG, Kirk PM, Kõljalg U, Kurtzman CP et al. . 2011. The Amsterdam Declaration on Fungal Nomenclature. IMA Fungus 2:105–112. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Hoog GS, Guarro J, Gené J, Figueras MJ (ed). 1995. Atlas of clinical fungi, 1st ed. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. [Google Scholar]
- 3.de Hoog GS, Guarro J, Gené J, Figueras MJ (ed). 2013. Atlas of clinical fungi, 3A ed. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands. [Google Scholar]
- 4.Schmalreck AF, Lackner M, Becker K, Fegeler W, Czaika V, Ulmer H, Lass-Flörl C. 2014. Phylogenetic relationships matter: antifungal susceptibility among clinically relevant yeasts. Antimicrob Agents Chemother 58:1575–1585. doi: 10.1128/AAC.01799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, Peterson SW, Varga J, Frisvad JC. 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud. Mycol. 70:159–183. doi: 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon-Chung KJ. 2012. Taxonomy of fungi causing mucormycosis and entomophthoramycosis (zygomycosis) and nomenclature of the disease: molecular mycologic perspectives. Clin Infect Dis 54(Suppl 1):S8–S15. doi: 10.1093/cid/cir781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hibbett SD, Ohman A, Kirk PM. 2009. Phenotypic variability: underlying mechanisms and limits do matter. New Phytol. 184:279–282. doi: 10.1111/j.1469-8137.2009.03042.x. [DOI] [PubMed] [Google Scholar]
- 8.O'Gorman CM, Fuller HT, Dyer PS. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 9.Houbraken J, Samson RA. 2011. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitt JI, Taylor JW. 2014. Aspergillus, its sexual states and the new International Code of Nomenclature. Mycologia. 106:1051–1062. doi: 10.3852/14-060. [DOI] [PubMed] [Google Scholar]
- 11.Sigler L, Sutton DA, Gibas CF, Summerbell RC, Noel RK, Iwen PC. 2010. Phialosimplex, a new anamorphic genus associated with infections in dogs and having phylogenetic affinity to the Trichocomaceae. Med. Mycol. 48:335–345. doi: 10.3109/13693780903225805. [DOI] [PubMed] [Google Scholar]
- 12.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O'Kelly MJ, van Oudenaarden A, Barton DB, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Smith F, Blomberg A, Durbin R, Louis EJ. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison CE, Hall C, Kowbel D, Welch J, Brem RB, Glass NL, Taylor JW. 2011. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc. Natl. Acad. Sci. U. S. A. 108:2831–2836. doi: 10.1073/pnas.1014971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neafsey DE, Barker BM, Sharpton TJ, Stajich JE, Park DJ, Whiston E, Hung CY, McMahan C, White J, Sykes S, Heiman D, Young S, Zeng Q, Abouelleil A, Aftuck L, Bessette D, Brown A, FitzGerald M, Lui A, Macdonald JP, Priest M, Orbach MJ, Galgiani JN, Kirkland TN, Cole GT, Birren BW, Henn MR, Taylor JW, Rounsley SD. 2010. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 20:938–946. doi: 10.1101/gr.103911.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira MM, Almeida-Paes R, Muniz MM, Gutierrez-Galhardo MC, Zancope-Oliviera RM. 2011. Phenotypic and molecular identification of Sporothrix isolates from an epidemic area of sporotrichosis in Brazil. Mycopathologia 172:257–267. doi: 10.1007/s11046-011-9437-3. [DOI] [PubMed] [Google Scholar]
- 16.Lackner M, de Hoog GS, Verweij PE, Najafzadeh MJ, Curfs-Breuker I, Klaassen CH, Meis JF. 2012. Species-specific antifungal susceptibility patterns of Scedosporium and Pseudallescheria. Antimicrob Agents Chemother 56:2635–2642. doi: 10.1128/AAC.05910-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner E, Jacobson DJ, Taylor JW. 2011. Genetic architecture of a reinforced, postmating, reproductive isolation barrier between Neurospora species indicates evolution via natural selection. PLoS Genet. 7:e1002204. doi: 10.1371/journal.pgen.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrone G, Stea G, Epifani F, Varga J, Frisvad JC, Samson RA. 2011. Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biol. 115:1138–1150. doi: 10.1016/j.funbio.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Varga J, Frisvad JC, Kocsubé S, Brankovics B, Tóth B, Szigeti G, Samson RA. 2011. New and revisited species in Aspergillus section Nigri. Stud. Mycol. 69:1–17. doi: 10.3114/sim.2011.69.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bensch K, Braun U, Groenewald JZ, Crous PW. 2012. The genus Cladosporium. Stud. Mycol. 72:1–401. doi: 10.3114/sim0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarver BAJ, Ward TJ, Gale LR, Broz K, Kistler HC, Aoki T, Nicholson P, Carter J, O'Donnell K. 2011. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet. Biol. 48:1096–1107. doi: 10.1016/j.fgb.2011.09.002. [DOI] [PubMed] [Google Scholar]