FIG 1.
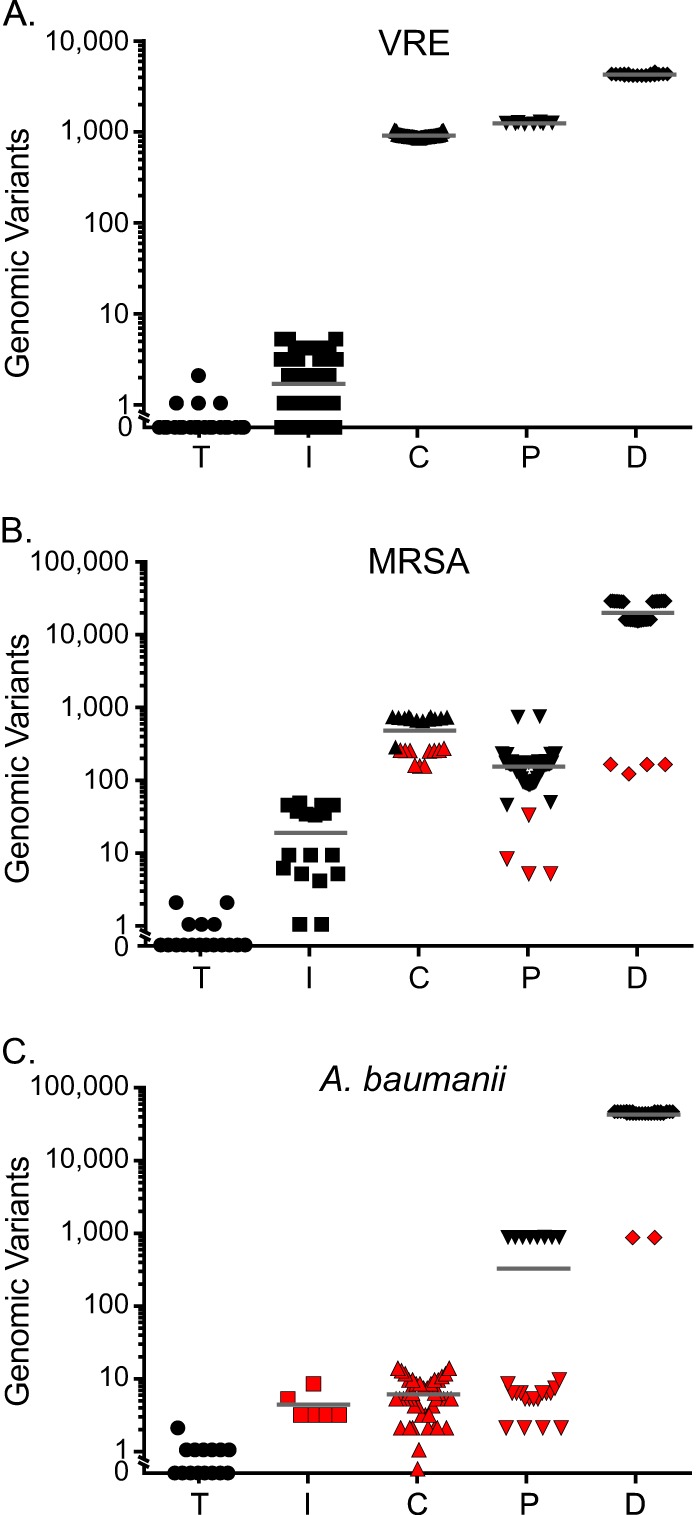
Numbers of disparate genomic polymorphisms detected among pairwise comparisons of technical replicates and different PFGE classification categories. Pairwise comparisons of isolates are stratified according to their status as whole-genome sequencing technical replicates (T) or classification by PFGE as “indistinguishable” (I), “closely related” (C), “possibly related” (P), or “different” (D). The numbers of genomic variants identified by whole-genome sequencing which distinguished isolate pairs are plotted along the y axis. Results are separately shown for VRE (A), MRSA (B), and A. baumannii (C). Red shading indicates a pairwise comparison with a count of genomic differences within 1 standard deviation of the mean observed in a different PFGE category for the same organism.
