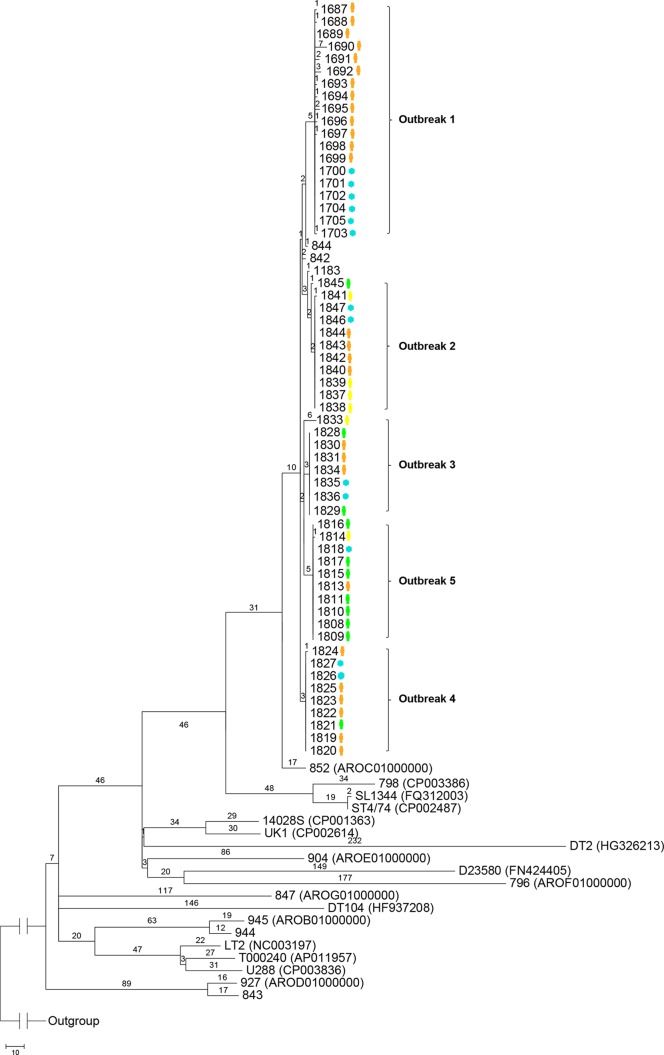
FIG 2.
Maximum-parsimony tree of S. Typhimurium genomes based on SNPs identified by mapping to the reference chromosome of S. Typhimurium LT2. Only SNPs in the “core” genome were included (27). The number on each branch is the number of SNP differences. Isolates representing each outbreak are demarcated with curly brackets followed by the outbreak numbers. The isolate source, either human (orange, epidemiologically confirmed; green, unknown epidemiological link; yellow, no epidemiological link) or environmental (blue), is noted next to the isolate number. In parentheses are the GenBank accession numbers of the publicly available genomes. The unit of the scale bar is the number of SNPs.