SUMMARY
Formation of the germline in an embryo marks a fresh round of reproductive potential, yet the developmental stage and location within the embryo where the primordial germ cells (PGCs) form differs wildly among species. In most animals, the germline is formed either by an inherited mechanism, in which maternal provisions within the oocyte drive localized germ-cell fate once acquired in the embryo, or an inductive mechanism that involves signaling between cells that directs germ-cell fate. The inherited mechanism has been widely studied in model organisms such as Drosophila melanogaster, Caenorhabditis elegans, Xenopus laevis, and Danio rerio. Given the rapid generation time and the effective adaptation for laboratory research of these organisms, it is not coincidental that research on these organisms has led the field in elucidating mechanisms for germline specification. The inductive mechanism, however, is less well understood and is studied primarily in the mouse (Mus musculus). In this review, we compare and contrast these two fundamental mechanisms for germline determination, beginning with the key molecular determinants that play a role in the formation of germ cells across all animal taxa. We next explore the current understanding of the inductive mechanism of germ-cell determination in mice, and evaluate the hypotheses for selective pressures on these contrasting mechanisms. We then discuss the hypothesis that the transition between these determination mechanisms, which has happened many times in phylogeny, is more of a continuum than a binary change. Finally, we propose an analogy between germline determination and sex determination in vertebrates—two of the milestones of reproduction and development—in which animals use contrasting strategies to activate similar pathways.
A hen is only an egg’s way of making another egg.
—Samuel Butler, 1885.
INTRODUCTION
The construction and successful implementation of a germline is an essential goal in development. Without a functional germline, sexually reproducing organisms1 cannot procreate, and the unique genetic composition of that individual is lost. The germline is a multigenerational, cellular lineage that includes the functional gametes, usually eggs and sperm, and all the precursor cells that lead directly to those fates. While many cells of an organism are necessary for the germline to develop and to be maintained, the germline lineage is distinct from the supporting or somatic cells of the organism. One could argue that the final lineage separation between the germline from its sister cells, which contribute to the soma, is the major milestone that determines the success or failure of a species. The germline contributes its genes to the next generation and thus in many ways has become immortal, whereas the somatic lineage ends with the adult.
The cycle of the germline is continuous. Following successful fertilization of functional gametes, the developing embryo will eventually make primordial germ cells (PGCs), a lineage that will commit largely if not exclusively to the germ cells. At some later point in development, the PGC population will expand through mitosis as germline stem cells (see Footnote 1) and eventually begin gametogenesis. Successful fertilization of the next generation completes the cycle. This scheme is true for the germline of most metazoans, although the details markedly vary between even closely related species. For excellent perspectives on diversity in reproduction, consider Birkhead (2000), Judson (2002).
The diversity in the germline continuum between species is undoubtedly dependent on several types of evolutionary mechanisms, including both adaptive and non-adaptive events. While natural selection has certainly dominated the thinking of phenotype evolution, other evolutionary forces must be integrated into the thinking of reproduction and are non-adaptive. These include DNA mutation (the ultimate source of variation in a population), DNA recombination (disperses mutations throughout the genome), and genetic drift (changes in the frequency of alleles in the population; see e.g., Gould and Lewontin, 1979; Lynch and Conery, 2003; Lynch, 2007). Each of these events is stochastic in nature, largely independent of the fitness properties of the individual (non-adaptive), and may have particularly important roles for diversification in the germline of a species population (Lynch, 2007). Non-adaptive evolutionary events likely also have different impacts on reproductive success and diversity depending on when the germ line is formed in development (e.g., Buss, 1987). Organisms that form their germline early in development, largely under maternal control, may have less individual and intra-species variation than an organism that relies on germline formation late in development, after many cellular divisions, or even in adulthood. For example, the germline of the fruit fly, Drosophila melanogaster, is formed after only 13 cell divisions, and is largely under the regulatory control of instruction left by the mother in the form of cytoplasmic deposits in the oocyte (e.g., Illmensee and Mahowald, 1974). The genome of the germline that forms in this animal will have relatively little time to vary from its somatic siblings. In contrast, the germline in plants; in the freshwater Cnidarian, Hydra; or the planarian, Schmidtea mediterranea, commit cells to the germline only once they are adults (Newmark et al., 2008; Steele, 2012). These cells have each undergone enormous numbers of divisions and have possibly been influenced by the environment differently over longer periods of biological time. Therefore, germline cells formed in these types of organisms may be genetically more variable than their siblings within the individual, and the germline then may report more variation acquired through this time. The core of these non-adaptive changes may then serve as the platform for adaptive changes that have led to the diverse germline phenotypes (Buss, 1987; Rosenberg, 2001; Lynch, 2007; Fernandez and Lynch, 2011).
In this review, we focus on the process of development that assigns the fate of PGCs in an embryo. We describe key molecular determinants of germline formation and their role in the two contrasting mechanisms of germline formation: inherited and inductive.2 We propose that a continuum exists between these two determination mechanisms, which may involve a series of changes that depend on the reproductive niche of the organism. The term reproduction niche was used recently (Bykova et al., 2012) to define the overall requirements that sexually mature plants have for successful reproduction: offspring production, dispersal, and germination. We use the term here similarly for animals, and add to it the intrinsic mechanisms used for mating, fertilization, specification of the PGCs, and development of gametes within the gonads, since clearly each of these features impinges on the overall ecological, temporal, developmental, and cellular space used to reproduce. We also draw upon an analogy between germline determination (inherited vs. inductive) and sex determination (environmental/temperature-dependent vs. genetic) inasmuch as both involve a conserved, core molecular basis that is controlled by rapidly evolving initiation mechanisms.
CORE GERMLINE DETERMINANTS—THE COMMON PATHWAY
Animals have relied on a conserved set of core genes to generate new germ cells in development. These include, but are not limited to, vasa, nanos, and piwi, and are found in all animals at some point in the construction or maintenance of a new germline. Many additional genes are necessary for an organism to uniquely construct a germ cell, although in many cases the identity of these genes remains uncertain. A molecular function, however, has been ascribed to each of the core germline gene products. The first, vasa, is a DEAD-box RNA helicase with strong sequence similarity to the translation factor eIF4A, and is thought to regulate the translation of mRNAs, perhaps even by selective interaction with mRNA sequences (Hay et al., 1988; Lasko and Ashburner, 1988; Linder et al., 1989; Liu et al., 2009; Gustafson and Wessel, 2010). Vasa is found in the germlines of all animals at some point in their development, and has been shown to be essential for PGC viability in at least in one sex or another in the many animals tested (e.g., Hay et al., 1988; Lasko and Ashburner, 1988; Tanaka et al., 2000; Raz, 2000; Medrano et al., 2012).
Perhaps prescient of a wider dependence of translational regulation in germline determination, other translational regulators also function selectively in the germline, including eIF4G3 in mouse sperm (Sun et al., 2010) and eIF4E isoforms during oogenesis in mammals and other tetrapods (e.g., Evsikov and Marín de Evsikova, 2009). Recently the initiation factor eIF4E-3 was found to be required for sperm development in Drosophila; meiotic chromosome segregation, cytokinesis, nuclear shaping, and individualization of sperm were each disrupted in its absence (Hernández et al., 2012). Despite the presence of other eIF4E isoforms (Drosophila has three), this loss-of-function effect on sperm development suggests that either germ cells have unique translational requirements served by this factor, or that the factor has essential roles in other cellular functions in addition to those of translation.
Vasa also appears to function outside of the germline in many animals, and is involved in regulating cell cycle activities. For example, vasa in Drosophila interacts with aubergine and spindle-E, two germline components of the Piwi-interacting RNA pathway, and recruits them for chromatin condensation during cell division (Pek and Kai, 2011a,b). In sea urchins, vasa associates with the mitotic spindle, where it is thought to regulate the translation of mRNAs important for the cell cycle, which may be limiting in large, rapidly dividing cells (e.g., blastomeres) of an early embryo (Yajima and Wessel, 2011). Most cells of the body divide quite well without Vasa protein, yet it appears to have an essential role in the cell cycle when it accumulates in somatic cells—even in tumor cells (Hashimoto et al., 2008; Janic et al., 2010; Wu and Ruvkun, 2010). Perhaps vasa imparts some unique regulation to the cell cycle that has been integrated within a different complex of checkpoints, and removal of vasa in those cells invokes activation of this modified checkpoint. The opposite perturbation, however, is not disruptive. Overexpression of Vasa protein does not cause any adverse phenotype in cell replication (e.g., Gustafson et al., 2011), so its function may normally be in excess (non-rate limiting) and have limited “off-target” effects. Further, ectopic Vasa is not sufficient for ectopic germ-cell formation (e.g., Gustafson et al., 2011).
Piwi, another conserved core germline gene, is a member of the argonaute family of small RNA-binding proteins. Originally found by a P-element insertion resulting in wimpy testes in Drosophila, it was identified as a gene required for stem cell maintenance in the Drosophila germline (Lin and Spradling, 1997). Later it was found to bind small (~30 nt) RNAs enriched in sequences from transposons, and was proposed to repress transposon function (Aravin et al., 2007; Brennecke et al., 2007). Such a model is intriguing since high transposon activity in the germline may be detrimental to the organism’s fitness (see Girard and Hannon, 2008) for a review of transposon control by small RNAs). It is now thought that piwi functions more widely both in terms of the cell types in which it functions and the processes it targets (Juliano et al., 2011; Juliano and Sneha, 2013, this issue), and that the generation of piRNAs appears to require both piwi and vasa in the process.
Many proteins are involved in piRNA biosynthesis, and a “ping-pong” (or “feed-forward loop”) mechanism was postulated for the generation of piRNAs in Drosophila (Brennecke et al., 2007; Gunawardane et al., 2007). This ping-pong amplification process is mediated by two Drosophila PIWI family proteins, aubergine (AUB) and argonaute3 (AGO3), which bind primarily to antisense primary piRNA and secondary sense piRNAs, respectively. Analysis of piRNAs in the germ cells of a vasa-deficient mouse showed that vasa plays an essential role in the early phase of the ping-pong amplification cycle. Kuramochi-Miyagawa et al. (2010) also suggest that vasa may play a role in the construction and/or function of intermitochondrial cement, and that Vasa is essential for the transfer of piRNA from the intermitochondrial cement to the so-called P-bodies, or processing bodies. The association of Vasa with small RNA pathways is consistent with the finding that its RNA-helicase activity is non-processive and maximal at 21 nt in length (Linder and Lasko, 2006; Sengoku et al., 2006), the average length of most miRNAs. This length is also close to the 26–31 nt of a piRNA, and even closer to the 21–24 nt piwi-associated small RNAs of Caenorhabditis elegans (Bagijn et al., 2012; Stower, 2012). Thus, Vasa may have important regulatory capabilities in multiple small RNA pathways, perhaps even by regulating miRNA association with target mRNAs through its helicase activity.
The third core element for germline determination, nanos, encodes an RNA-binding protein that interacts with a ubiquitous Pumilio protein (Irish et al., 1989; Asaoka-Taguchi et al., 1999). The nascent Nanos/Pumilio complex binds to specific sequences in the 3′-untranslated region (UTR) of messages that contain a Nanos response element (NRE) and recruits proteins that feed back onto the 5′-UTR to halt translation initiation in Drosophila (Forbes and Lehmann, 1998; Sonoda and Wharton, 1999; Gupta et al., 2009). This complex may also recruit a de-adenylase complex that removes the 3′-UTR polyadenylated tail, thus causing specific mRNA degradation (Suzuki et al., 2010, 2012). The activity of Nanos may therefore have multiple functions in germ-cell formation, including slowing the cell cycle early in PGC formation through repression of cyclin B activity (Kadyrova et al., 2007). Pumilio is more generally present throughout an embryo, but cells have acquired multiple mechanisms to restrict the expression of nanos to the germline (Sato et al., 2007). Nanos protein appears to be “toxic” outside of the germline, that is, Nanos mis-expression can repress cell cycling in somatic cells as well as lead to misappropriation of cell fates, presumably through its interaction with Pumilio on mRNAs that regulates the cell cycle (Lai et al., 2012). Germ cells lacking nanos usually undergo apoptosis through the translation and activation of hid, an apoptotic initiator whose mRNA is normally repressed by the Nanos/Pumilio complex in the germline (Sato et al., 2007).
Each of these core, germline determination genes is used by all metazoans whose germlines have been studied—yet when, where, and how they are utilized during the developmental program of the germline lineage differs enormously from species to species. Part of these differences in timing may reflect a modular function of the core germline genes, and the many additional gene products used to establish a germline. Another difference is the diverse reproductive strategies used by each embryo to establish its germline.
CONTRASTING STRATEGIES OF GERMLINE DETERMINATION
The many divergent strategies for germline determination are often clumped into three main types: adult multi-potent stem cell-derived, inherited, and inductive.
Multipotency
Many metazoans, as well as flowering plants, retain cells with multipotentiality throughout adulthood, capable of differentiating into diverse cell types. Flowering plants do not contain a distinct germline in embryos, but they do maintain populations of undifferentiated stem cells that continuously produce both vegetative tissues and organs that lead to formation of the germline in the adult. Cells within these tissues can switch to the production of reproductive organs containing diploid sporogenous cells (gametophytic cells), which will give rise to the haploid male and female gametes. Thus, germ cell precursors can form anew in these adults. Although the mechanisms of this adult germline determination process are not resolved in flowering plants, the mechanisms do not contain the same core germline genes as seen in animals (see e.g., Berger and Twell, 2011 for review of germline specification in plants). Similarly, stem cells in many metazoans have the potential to become a broad palette of somatic cells (as well as germ cells), and are established early in development. In the Cnidarian Hydra magnipapillata and in the Lophotrochozoan flatworm Schmidtea mediterranea, specific stem cells are established in development and are maintained in adulthood. These so-called interstitial stem cells (I-cell) of H. magnipapillata and the neoblasts of S. mediterranea are capable of differentiating into diverse cell types, including germ cells.
Planarians have the ability to rapidly regenerate regions of their body. This regeneration has been attributed to the presence of totipotent neoblasts (Baguñá, 1981). The neoblast cells contain round, electron-dense chromatoid bodies that are similar to germ granules in other organisms and are maintained in germ cells after differentiation (Pederson, 1959; Morita, 1967; Coward, 1974; Hori, 1982). Experiments on the planarian Dugesia japonica have shown that vasa-related gene products are present in the chromatoid body and may play a role in neoblast totipotency (Shibata et al., 1999). A nanos-related gene, djnos, is found within a subpopulation of the neoblasts that produce germ cells during sexualization (Sato et al., 2006). Macrostomum lugano expresses homologues of vasa (macvasa) and piwi (macpiwi) in the gonads and somatic stem cells of males and females (Pfister et al., 2007,2008). In S. mediterranea, neoblasts are the only dividing cell of the adult and are responsible for all cell types of the organism (Wang et al., 2007; Forsthoefel et al., 2012). In addition to the somatic cells, these adult stem cells contribute to the germline, and when they do so, they appear to use the same core germline gene set as used in more complex animals, including vertebrates (Mochizuki et al., 2000, 2001).
Inheritance
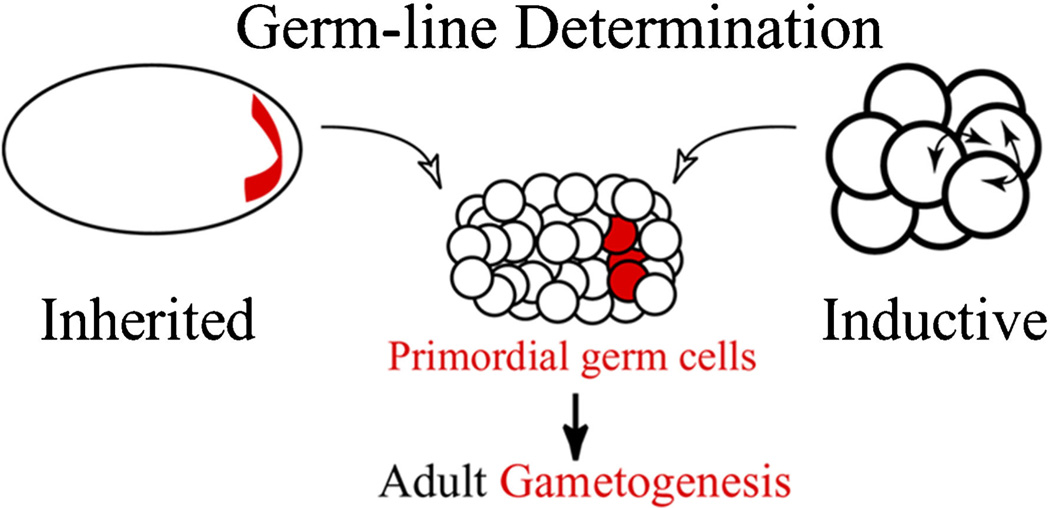
Inheritance-based germline determination refers to a mechanism whereby maternal products accumulate in one region of the egg or early embryo, and the cells that acquire that material begin a process of germline determination (Fig. 1). This developmental mechanism is present in many invertebrates and vertebrates, and is most thoroughly described in the model organisms D. melanogaster, C. elegans, Danio rerio (zebrafish), and the frog Xenopus laevis (e.g., Seydoux and Braun, 2006; Ewen-Campen et al., 2010). In these animals, the core germline gene set (vasa, nanos, and piwi) are key “germline determinants,” and are present within distinct cellular structures, such as pole plasm (Drosophila, Zebrafish), and germ plasm (Xenopus), which are morphologically different but serve a similar function.
Figure 1.
Inherited versus inductive germline determination. Embryos have two different strategies to determine their primordial germ cells. Inherited mechanisms rely largely on localized maternal determinants that, when acquired by cells of the embryo, assign the fate of a germline to those cells. In contrast, inductive mechanisms rely on intercellular inductive interactions that are interpreted by the cells to acquire a germline fate. Although each mechanism is distinct in principle, they converge on activation of a similar core gene set for maintenance and differentiation of germ cells. These elements include Vasa, Nanos, and Piwi (shown in red).
In Drosophila, the oskar gene controls assembly of the germ plasm and determines the number of precursors formed at the posterior end of the embryo (Ephrussi et al., 1991). Downstream interaction of oskar with vasa is required for the assembly of polar granules (Breitwieser et al., 1996). Tudor, another gene essential for polar granule assembly (Thomson and Lasko, 2004), is downstream of vasa in the pathway for germ-cell fate. Formation of germ cells at an ectopic site in Drosophila requires these three genes only (Ephrussi and Lehmann, 1992; Smith et al., 1992). Nanos RNA also localizes to the germ plasm (Nakamura et al., 1996) and acts downstream of vasa, oskar, and tudor to regulate posterior pattern formation of the Drosophila embryo (Lehmann and Nüsslein-Volhard, 1991; Wang and Lehmann, 1991). A gene with a function similar to oskar (Ewen-Campen et al., 2010), and the first gene to be identified as essential for germ plasm organization in vertebrates (Bontems et al., 2009), is bucky ball. In zebrafish, it regulates the formation of the Balbiani body (also known as the mitochondrial cloud in Xenopus), an aggregation of cellular organelles transiently found in the oocytes of all examined vertebrates using inherited mechanisms of germline determination (Billett and Adam, 1976). Bucky ball also determines the animal-vegetal axis in zebrafish through the localization of appropriate transcripts, and is thus responsible for the polarity of the oocyte and early embryo (Marlow and Mullins, 2008). Initially, vasa is maternally expressed throughout the zebrafish embryo (Braat et al., 2000), but loss of vasa through morpholino-mediated knockdown does not affect formation of germ cells (Knaut et al., 2000; Braat et al., 2001). A nanos-like gene (nanos1) localizes to the germ plasm and is essential for migration and survival of the germ cells in the embryo (Koprunner et al., 2001); in the adult fish it is required for production of oocytes (Draper et al., 2007).
The structures mentioned above are enriched within a small region of the egg or early embryo, and the cells that acquire these structures are directed to a germline fate (see also Gao and Arkov, 2012). A major research question in the field is what functions are present in those cellular structures that direct naïve cells to a germ-cell fate. Clearly, just adding vasa, nanos, or piwi to a cell is insufficient to direct that cell to a germline fate (see e.g., Gustafson et al., 2011; Lai et al., 2012). These factors are necessary, but not sufficient, for germline determination. Instead, a large combination of functions that broadly changes the cellular activities is required, including alterations in chromatin structure, translational changes, mRNA and protein turnover, and reconfiguration in the cell-surface proteome.
Recently, the germ plasm of an early embryo has been shown to be sufficient for germ-cell formation in a vertebrate. Tada et al. (2012) transplanted eGFP-labeled germ plasm from an eight-cell embryo into ectopic sites of a stage-matched Xenopus embryo, leading to PGC formation in a distal site. These ectopic PGCs became functional germ cells, both sperm and eggs, indicating that the germ plasm of frogs is sufficient as an inherited mechanism for germ-cell fate determination. Moreover, this emphasizes the dramatic similarity to what was learned by similar approaches in Drosophila (Illmensee and Mahowald, 1974; Mahowald, 2001) and suggests similar inheritance mechanisms. Not only was the germ plasm capable of re-directing the fates of the recipient cells to the germline, but the recipient cells were also capable of “reading” the germ plasm and responding appropriately. Although the newly formed PGCs did not migrate to the developing gonads on their own, they were able to integrate into the somatic cell matrix and develop into functional gametes if transplanted near the developing gonad site. This result also supports the model that germ plasm is not simply a marker of the germline, for example, a convenient tool for investigators, but a functional element that actively regulates germline determination.
Recent results in C. elegans suggest that P-granule segregation, long thought to be an essential element of germline determination, actually is not essential for germline determination (Gallo et al., 2010; Voronina, 2012; see also Strome et al., 1995; Hird et al., 1996). Genetic-based disruption of the P-granules (by mutation of a phosphatase enzyme, pptr) causes dispersion of the P-granules and an absence of segregation during early cleavage divisions, although normal germline determination and reformation of P-granules occurs later in development and the animals are fertile. Clearly, more than the core elements of the germline determination pathway are needed for this cell fate. This result also underscores the importance of identifying the contents of localized materials, their mechanisms of action, and accessory factors essential for this function. The above result may also be a clue to how some embryos can restore germline stem cell function following disruption or removal of their initial assignment. Such a “recovery” of PGCs has been seen in ascidians (Takamura et al., 2002) and in sea urchins (Voronina et al., 2008).
Inductive Determination
A third major mechanism of germline determination is referred to as inductive or induction-based. Animals using inductive germline determination mechanisms have no predefined germ plasm or localized determinants in the embryo that directly lead to a germline fate. Instead, cells in the embryo rely on positional information and intercellular communication to ascribe this fate decision (Fig. 1; McLaren and Lawson, 2005). Without specific inductive signaling, cells are directed instead to continue towards a somatic cell fate. The process of induction may even involve a diversion from the somatic cell fate in certain cells, thereby restricting them to a germline fate. These developmental processes occur later in development than for the inherited mechanism of germ-cell fate, and PGCs are therefore established later in development. The inductive developmental strategy appears to be the most ancient mechanism used by animals, and is present widely in animal taxa. It is believed that the inherited, germ plasm-driven mechanisms have independently evolved several times throughout the phylogeny (Extavour and Akam, 2003; Ewen-Campen et al., 2010), perhaps resulting in the vast differences in structures of these localized determinants.
The mechanisms for inductive determination are not as well understood as the inherited mechanism. Even though they are distinct strategies and are seen in different species, the same core germline gene set is still employed (Ewen-Campen et al., 2010). Additional layers of complexity may also be involved because of the inductive signaling necessary for this mechanism and because it may generally rely more on mechanisms of repression and/or redirection of somatic cell fate. In contrast, cells in the early embryos of organisms using inherited mechanisms show all possible cell fate potentials at the time of inheritance, and have the task of maintaining those fates for later development. Inductive instruction may necessarily rely on cellular communication, leading to new gene expression and chromatin modifications that effectively repress an otherwise budding somatic fate within the cell. The best-studied example of inductive specification of the germline is in the mouse, where extensive classic experimental embryology is partnered with a contemporary molecular approach to cellular interactions and chromatin modifications.
Mice differ from organisms using inherited mechanisms (Drosophila, Xenopus, etc.) in that the embryos show no allocation of the germline during early cleavage stages (see Durcova-Hills and Capel, 2008). Instead, the PGCs in mice are derived from the epiblast during gastrulation as a result of signals from the surrounding extraembryonic ectoderm and visceral endoderm (Tam and Zhou, 1996; McLaren and Lawson, 2005, Hayashi et al., 2007). The inductive nature of germline determination in this organism was originally explored by cell transplants: cells from the distal epiblast of a mouse embryo were transplanted to the proximal region of the epiblast, where they turned into PGCs, a fate distinct from their previous location (Tam and Zhou, 1996). This suggests that it is location and communication, not acquisition of stored materials (e.g., germ plasm), that determines cell fate in this animal. It does not, however, exclude the possibility that only certain cells of the epiblast—perhaps having acquired some unique maternal constituents (vis-à-vis inherited specification)—are capable of responding to the given intercellular signal (e.g., inductive; see also Fig. 2).
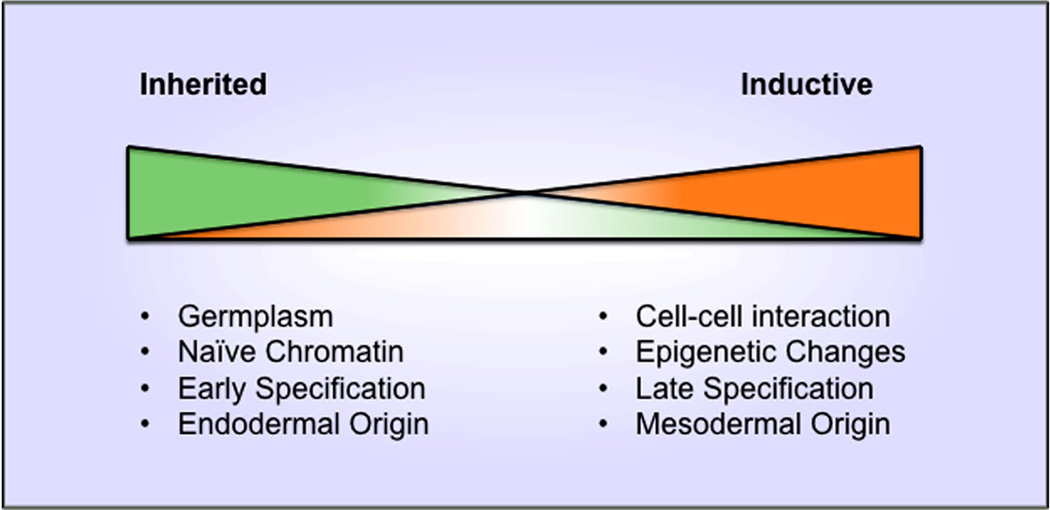
Figure 2.
A continuum of mechanisms used for germ-cell specification. At the extremes are the consensus mechanisms of inherited and inductive germline determination. These conclusions are bases on the very few embryos that have been studied in detail. Each of the cellular and molecular events is likely used to differing degrees in a species, forming a continuum of mechanisms across phyla. For instance, some embryos using inherited germline determination may actually specify their germline later, requiring more chromatin modifications than the extreme case.
The PGCs that form in mice soon show characteristic staining with tissue non-specific alkaline phosphate (TNAP; Chiquoine, 1954), the original germline marker found in many vertebrates. Ginsburg et al. (1990) show that about eight such cells cluster together at a location posterior to the definitive primitive streak, and by 7.5 days post-coitum (dpc), this cluster grows into the founder pool of 40 PGCs (McLaren and Lawson, 2005). Anderson et al. (2000) found that a truncated oct4 promoter, a gene later associated with pluripotency, steminess, and induction of pluripotency stem cells from fibroblasts (iPS cells; Takahashi and Yamanaka, 2006; see also Yeom et al., 1996), is able to drive GFP expression selectively in PGCs. This discovery has been particularly useful in determining mechanisms of migration and the fates these cells acquire in the event of changes in the environment or of the genes normally regulating their fate. Thus, PGCs can now be visualized by TNAP and GFP expression, although this approach does not elucidate the molecular mechanisms of germline determination in the animal since it is a later event resulting from initial specification mechanisms.
How is inductive specification of the germline achieved? The PGC markers previously used in other organisms (nanos, vasa, and piwi) do not accumulate until later in development in the mouse, and do not reveal any of the early specification programs required for inductive mechanisms. It is as if the mechanism of inherited specification (e.g., in Drosophila and Xenopus) has expedited the processes, whereas in the mouse and other mammals, these same essential genes are only invoked later in the germline developmental process (Fig. 2).
Many labs have made significant progress in the past several years into the mechanisms of inductive germline specification in the mouse. As a result, we now also have an opportunity to comparatively analyze these mechanisms in a wide range of species displaying this ancient mechanism of germ-cell determination. Several gene functions have been discovered recently that together support the hypothesis that the inductive mechanism of germline determination emphasizes broad genomic regulators that both repress somatic fates and specify a germ cell.
THE QUEST FOR A MOLECULAR MECHANISM OF INDUCTIVE GERMLINE DETERMINATION
A decade ago, Saitou et al. (2002) set out to uncover the molecular program by which mouse PGCs are set aside from their somatic neighbors. The team’s approach was to interrogate mRNAs from individual cells separated from the posterior epiblast where the PGCs form. They made complementary DNA (cDNA) libraries from individual cells and tested if mRNAs accumulated with an expression pattern that they predicted to be limited to PGCs. Their approach was creative and logical, and used an iterative positive/negative selection of PCR products from each of the individual-cell cDNA libraries. They started with bone morphogenetic protein-4 (bmp4). When the extraembryonic tissue of the mouse is mutant for bmp4, no germ cells form in the embryo even if the embryonic tissue is wild-type. Conversely, wild-type extraembryonic tissue supports formation of germ cells in an epiblast that is mutant for bmp4 (Lawson et al., 1999). This result showed that Bmp4 signaling from the extraembryonic ectoderm lays the foundation for the founder pool of PGCs.
It was later confirmed that the germ-cell fate is a direct consequence of bmp4 signaling (Ohinata et al., 2009). Cells of the allantoic mesoderm do not express bmp4, allowing this gene to be used as a negative marker in the initial screen for candidate PGCs. Since the candidate PGCs are found in the region of the epiblast adjacent to the extraembryonic material, it was difficult to determine whether a cell was from the epiblast or the extraembryonic ectoderm. The “Saitou Screen” used the absence of bmp4 as a preliminary indication that the selected cells were from the epiblast. TNAP served as a positive marker for cells that constitute the germline. Finally, the screen used hoxb1 as a negative marker for PGCs, as it is expressed in somatic cells of the epiblast, but not in the germline. Ten of 83 cells from the proximal epiblast stood out from this positive/negative interrogation as nominees for a PGC fate.
The candidate PGCs were then further analyzed for genes that may be unique to the cell at the time of cell fate specification, and the test produced two gene products. Fragilis (iftm3 in Tanaka and Matsui, 2002), a newly discovered member of the interferon-(IFN)-inducible transmembrane protein family, was the first gene to mark germ-cell competence. It can be detected also in many different adult and embryonic tissues, which suggests that it has a broader role than just in germ-cell determination (Saitou et al., 2002). For several years, the presence of fragilis in pre-migratory PGCs and its conservation in mammals led researchers to believe that it must have some role in PGC determination (Lange et al., 2003; Tanaka et al., 2005; Tam and Loebel, 2007). Once the gene function was tested by knockout mutations, however, the animals did not show any signs of developmental problems, inviability, or infertility (Lange et al., 2008). Thus, even though fragilis is the earliest of known germline marker expressed in the mouse, its function in this context is still unknown. In the least, it may have functional overlap with other genes within the cell that impart functions that are not yet appreciated. Until this is resolved, it still serves a useful purpose as an additional marker of the germline, as both fragilis and TNAP are not required for PGC development or migration (MacGregor et al., 1995).
The second gene emanating from the “Saitou Screen” was stella (PGC7/Dppa3 in Sato et al., 2002), a maternal effect gene that maintains DNA methylation and is involved in epigenetic reprogramming post-fertilization (Nakamura et al., 2007). Recent work shows that it prevents the conversion of 5-methylcytosine to 5-hydroxymethylcytosine in the maternal genome (Nakamura et al., 2012). It is expressed later than fragilis, and is a marker of lineage-restriction, although it too is dispensable for germ-cell determination and development (Payer et al., 2003; Bortvin et al., 2004). The cells expressing fragilis and stella in the “Saitou screen” showed clear suppression of hox genes and high expression of two mesodermal markers (T, the mouse homologue of Brachyury, and Fgf8). This provides molecular evidence that PGCs arise from cells that are originally destined for a somatic-mesodermal fate, an important feature of inductive germline determination. In support of this conclusion, the group never detected stella in cells lacking fragilis in vivo (Saitou et al., 2002). Interestingly, epiblast cells induced by the extraembryonic ectoderm in vitro show expression of fragilis but not stella. This finding opened up the possibility of an additional signal—one that would lead to lineage-restriction and the final stages of PGC specification.
A breakthrough came in January 2005, when Vincent et al. (2005) learned that a DNA-binding transcriptional repressor known as B-lymphocyte induced maturation protein 1 (blimp1) is necessary for the formation and specification of PGCs. Blimp1 was isolated as a positive regulatory domain I-binding factor (prdm1), a novel repressor of beta-interferon expression (Keller and Maniatis, 1991). It has since been shown to carry out a number of functions ranging from forelimb development to tumor suppression (see Bikoff et al., 2009; John and Garrett-Sinha, 2008, and Martins and Calame, 2008 for review of functions). The gene has an established role in PGCs: blimp1-positive PGCs are detected at 7–8 dpc. These cells migrate to the gonads at 11–12 dpc, and stop expressing blimp1 at 13 dpc (Chang and Calame, 2002). Vincent et al. (2005) originally aimed to test the role of blimp1 in establishing the antero-posterior axis, and went on to show various abnormalities in loss of function (LOF) mutants. Blimp1 LOF embryos fail to survive past 10.5 dpc as a result of defects in their branchial arches, severe hemorrhaging, and even placental defects. Of great importance here is the finding that blimp1 mutants show a dosage-specific decrease in PGCs to a point where homozygous mutants lack PGCs altogether. This work has provided researchers with blimp1 as a candidate gene that might be responsible for PGC lineage-specification in mice.
THE ASCENT OF blimp1 AND EPIGENETICS IN INDUCTIVE GERMLINE DETERMINATION
Ohinata et al. (2005) found approximately six epiblast cells showing expression of blimp1 as early as 6.25 dpc. At 7.5 dpc the number of cells increases to a founder pool of 40 cells, which in turn express stella and then appear to only demonstrate PGC fates. It is now clear that the initial six cells are the earliest members of the germ-cell lineage, but they are by no means the only ancestors (Lawson and Hage, 1994; Tam and Zhou, 1996; reviewed in McLaren and Lawson, 2005). A lineage-tracing experiment involving a Cre-LoxP system helped to establish that blimp1-positive cells expressing stella are restricted to a germ-cell fate (Ohinata et al., 2005). The above study corroborated the previous finding (Vincent et al., 2005) that blimp1-negative embryos lack PGCs. cDNA libraries (similar to those from the “Saitou Screen”) of blimp1-deficient cells reveal that the few stella-positive mutant cells seem to be perturbed in several aspects compared to wild-type PGCs. These cells show inconsistent expression of the hox genes, fail to proliferate, and it was speculated that they eventually undergo apoptosis or adopted a somatic cell fate that was assigned to them prior to initiation of the germline-specification program (Ohinata et al., 2005). It seems clear that blimp1 is indeed a critical determinant of the germ-cell lineage.
Blimp1 is involved in a transcriptional regulatory complex that plays a unique role in the mouse germ-cell lineage (Ancelin et al., 2006), and has been implicated as an epigenetic repressor (Lin et al., 1997; Ren et al., 1999; Piskurich et al., 2000; reviewed in Kouzarides, 2002). To test this hypothesis in PGCs (see Bikoff et al., 2009), Ancelin et al. (2006) looked for histone methyltransferase activity that was exclusive to the PR domain, a derivative of the SET (suv39, enhancer of zeste, trithorax) protein domain (Ohinata et al., 2005). A myc-tagged blimp1 immunoprecipitate was used to conduct a histone methyltransferase activity assay. Histones H3 and H2A stood out as candidate target proteins in the assay, and blimp1 led to their methylation on arginine 3 (H2A/H4R3me2) of the conserved five amino acid N-terminus. The group speculated that since PR-SET domains are known to methylate lysine, not arginine, there must instead be another protein with histone arginine methyltransferase activity present in the immunoprecipitate. Indeed, protein arginine methyltransferase-5 (prmt5), which functions in dimethylation of histone arginine residues (Branscombe et al., 2001; Pal et al., 2004), was found to associate with blimp1 in vitro and in vivo.
Prmt5 is a maternally inherited gene that is up-regulated in the cytoplasm during the derivation of embryonic stem cells (ESCs) and interacts with mep50 in order to maintain ESC pluripotency (Tee et al., 2010). Mep50 (methylosome, WD-repeat containing protein) is important for methylosome activity, specifically methylating predeposition histones H2A/H2A.X-F and H4 and the histone chaperone nucleoplasmin on a conserved motif (GRGXK; Friesen et al., 2011; Wilczek et al., 2011). The Blimp1-Prmt5-Mep50 complex was found to be responsible for high levels of H2A/H4 R3 methylation when present in the nucleus at 8.5 dpc. The presence of the complex within the nucleus correlated with down-regulation of the dhx38, a gene whose ortholog in C. elegans has been implicated in germline development (Graham and Kimble, 1993). Dhx38 encodes a DEAH box-containing RNA helicase activity (Ancelin et al., 2006), and its gene has four GGGAAAG motifs corresponding to a blimp1 binding site (Kuo and Calame, 2004). The sequence present in exon 11 of the dhx38 gene is the target of the Blimp1/Prmt5 complex. dhx38 expression is up-regulated once Blimp1-Prtm5 moves from the nucleus to the cytoplasm (at 11.5 dpc), and the PGCs go into mitotic/meiotic arrest by 12.5 dpc. High levels of prmt5 and dhx38 are also detected when similar tests are performed in ESCs, embryonic germ cells, and P19 mouse carcinoma cells, all of which are pluripotent (Ancelin et al., 2006). Not surprisingly, blimp1 is absent in these cells. Thus, the spatial and temporal regulation of H2A/H4 R3 dimethylation in vivo was proposed to play a crucial role in PGC specification, providing a foundational mechanism for this complex process.
Given these findings, blimp1 was postulated as a gene responsible for repression of the somatic cell program and maintenance of early germ-cell characteristics (Hayashi et al., 2007). In quick order, blimp1 was reported to repress nearly all the genes normally down-regulated in PGCs as compared to the neighboring somatic cells (Kurimoto et al., 2008). Lin28, a negative regulator of the let-7 microRNA (miRNA), is required for PGC development, and overexpression of lin28 promotes the formation of stella-positive cells and PGCs in vitro and in chimeric embryos, respectively (West et al., 2009; reviewed in Matzuk, 2009). As a target of let-7 miRNA (Ohinata et al., 2005), blimp1 was shown to rescue the effects of lin28 knockdown by RNA interference (RNAi), thereby establishing lin28 as an upstream component of the PGC development pathway (West et al., 2009). Importantly, new technology emerged that allowed blimp1 and stella expression to be analyzed both in vitro and in vivo. A double-transgenic line was used to express membrane-targeted Venus (mVenus), a brighter version of yellow fluorescent protein (YFP), and enhanced cyan fluorescent protein (eCFP) as reporters for blimp1 and dppa3 (stella) expression, respectively (Ohinata et al., 2008). One of the main problems in visualizing PGCs in vivo was that the cells are often unable to sufficiently up-regulate a single transgenic reporter because of the variable chromatin states of its genome. The double-transgenic approach not only bypassed this problem and provided a non-invasive method to visualize PGC development in vivo, but also provided researchers with an opportunity to induce expression of germ cells in vitro with temporally coordinated blimp1 and stella expression.
Although blimp1 and Prmt5 form a complex in the process of germline determination, their individual actions appear to be antagonistic in the grand scheme of germline determination. First, it is known that blimp1 and prmt5 are respectively up-regulated and down-regulated in ESCs (Ancelin et al., 2006; Tee et al., 2010). Second, evidence suggests involvement of reprogramming (de-differentiation) in PGCs to pluripotent embryonic germ cells. When exposed to the endogenous signaling molecule fibroblast growth factor 2 (FGF-2), PGCs down-regulate Blimp1 (Durcova-Hills et al., 2008). Subsequently, three known targets of Blimp1-dhx38 (Ancelin et al., 2006), c-myc (Lin et al., 1997), and klf-4 (Durcova-Hills et al., 2008)— are all up-regulated in tandem with the loss of blimp1 in these cultured PGCs. Prmt5 is retained in embryonic germ cells, and takes on a cytoplasmic role similar to that in naturally occurring ESCs. Induction of blimp1 in mouse embryonic fibroblasts leads to partial reprogramming of somatic cells, whereas induction of prmt5, klf4, and oct3/4 gives somatic cells the potential to differentiate into all three germ layers in a manner comparable with ESCs (Nagamatsu et al., 2011). Thus, prmt5 appears to be a gene responsible for maintaining the pluripotency of cells, whereas blimp1 imposes some restriction on the fate of mesodermal cells during PGC specification and formation. Bao et al. (2012) have confirmed that blimp1 is not required for the derivation of ESCs from the epiblast or for their maintenance.
A SIBLING IS FOUND
The identification of prdm14, a close relative of blimp1/prdm1, defines a novel pathway for ensuring the development of the germ-cell lineage (Yamaji et al., 2008; reviewed in Bikoff and Robertson, 2008). Expression of prdm14 is seen by 6.25 dpc (Yabuta et al., 2006), preceding expression of stella, and was found to be critical for germ-cell development, via repression of the somatic mesodermal program, but is not required for embryonic development. The proline-rich sequence in Prdm1/Blimp1 might contribute to its array of functions in cell types outside the germline (Bikoff and Robertson, 2008), thus making it essential for embryonic development and viability (Vincent et al., 2005), but also provides functional overlap to prdm14. Prdm14 is essential for the derivation of embryonic germ cells from PGCs via the reacquisition of pluripotency, and has a defined role within the PGC-specification pathway. Although initial activation in the germline is blimp1-independent, the “master PGC regulator” controls prdm14 up-regulation and maintenance. The bmp4-smad1 pathway ultimately controls expression of both these major transcriptional regulators (see Fig. 3, inlay). Although the details remain murky, involvement of the bmp4 pathway could be predicted given that the gene is essential for mesoderm formation (Winnier et al., 1995) and that PGCs arise from cells otherwise destined for a mesodermal fate (Saitou et al., 2002). Prdm14 is also involved in somatic cell reprogramming, although, much like with blimp1, cells such as embryonic fibroblasts expressing the gene do not become pluripotent (Nagamatsu et al., 2011). In humans, prdm14 is required, but not solely sufficient, for maintaining self-renewal of ESCs, although it does induce suppression of genes that keep them in an undifferentiated state (Tsuneyoshi et al., 2008).
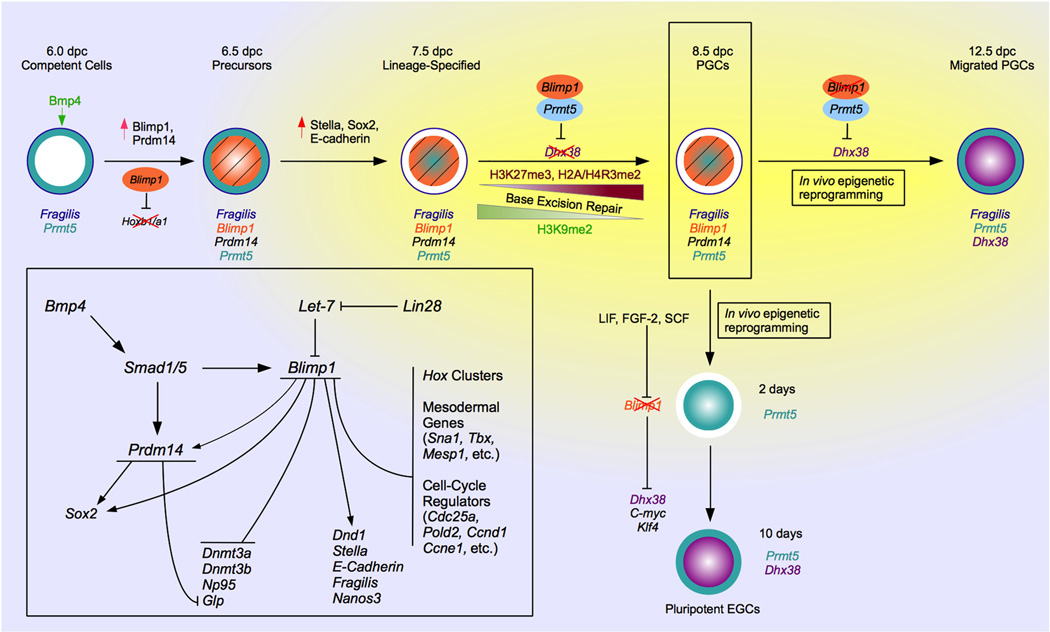
Figure 3.
A molecular mechanism for inductive germ-cell specification and beyond. The process of PGC specification begins with the expression of fragilis in the cell membrane. The Bmp4 path way triggers expression of blimp1 and prdm14, the master regulators of the specification process. The blimp1/Prmt5 complex suppresses expression of dhx38; coupled with epigenetic changes, this gives rise to PGCs at 8.5 dpc. From here, cells undergo reprogramming in vivo during migration to the genital ridge. The cells can also be converted to pluripotent embryonic germ cells though in vitro reprogramming. Note that certain processes (loss of blimp1, translocation of Prmt5 to the cytoplasm, expression of dhx38, and other blimp1 targets in the nucleus) are conserved across both the processes. Inlay: Gene pathway showing blimp1 and prdm14 as key players in the conditional germline determination process. Adapted from Saitou and Yamaji (2010).
The proposed model from these findings (Fig. 3; see also Arnold and Robertson, 2009; Saitou, 2009; Saitou and Yamaji, 2010) suggests that a select number of cells at the posterior end of the mouse epiblast express prdm1 and prdm14 in response to bmp4 signaling from the extraembryonic ectoderm. Cells that do not express these genes instead express the hox genes, and head toward a somatic mesodermal fate, prdm-positive cells, however, show up-regulation of stella, sox2, and other key genes involved in the germline, and down-regulation of the hox genes. Chromatin modification in these cells begins with the Blimp1/Prmt5 complex methylating H2A and H4 on arginine 3 (H2A/H4 R3Me2). Specification is completed following increased trimethylation of lysine 27 and decreased dimethylation of lysine 9 on histone H3 (H3K27me3 and H3K9Me2, respectively). These final modifications in nascent PGCs are not linked to the Blimp1/Prmt5 complex, but have alternate mechanisms (Seki et al., 2007; Ohinata et al., 2009; see also Kota and Feil, 2010; Gillich and Hayashi, 2011). For example, it is now believed that DNA demethylation in mouse PGCs is a result of single-stranded DNA breaks and a process of base excision repair (Hajkova et al., 2010; reviewed in Surani and Hajkova, 2010). During in vitro reprogramming of PGCs to ESCs, blimp1 is down-regulated so that Prmt5 is left in the nucleus, and is eventually translocated to the cytoplasm (Durcova-Hills et al., 2008). The genes targeted by blimp1 are up-regulated, giving rise to self-renewing pluripotent cells. The transition from PGC to embryonic germ cell is accompanied by expression of stat3, klf4, and c-myc over a course of approximately 10 days.
Does blimp1 activity intersect with fragilis and stella? What role do these two play, given that they have been shown to be dispensable for PGC specification and fertility in the mouse? Fragilis is an interferon-inducible protein located in the plasma membrane at cell-to-cell contact sites (Saitou et al., 2002). Since blimp1 was isolated via its ability to bind and repress a beta-interferon promoter, perhaps a connection exists between expression of the two genes at the onset of PGC specification. It is also unknown if the Blimp1/Prmt5 complex interacts with stella at a molecular level, even though they are both present in the nucleus around 7.5 dpc. The first study of blimp1 knockouts (Vincent et al., 2005) showed that the PGCs present in a few of the mutants failed to migrate to the genital ridge. Yet, no studies have pursued this observation further, probably because of the embryonic-lethal nature of the mutation. Nanos3, a member of the nanos family of genes described previously, is found in migrating PGCs (Tsuda et al., 2003), so a nanos3::cre-recombinase gene construct, for example, could be used to excise a floxed blimp1 gene in migrating PGCs in order to determine whether or not blimp1 is essential for this process. Such a conditional, tissue-specific knockout of blimp1 might provide greater insight into the role of the master-regulator in this key process that takes place post-specification.
Mouse epiblast cells experience major transcriptional and epigenetic changes from pre- to post-implantation development. These changes include DNA methylation, chromatin compaction, random X-chromosome inactivation, and silencing of pluripotency genes (Monk and Harper, 1979; Ahmed et al., 2010; Borgel et al., 2010; Miyanari and Torres-Padilla, 2012). Reversal of many of these changes occurs uniquely within the PGCs following their specification, and appears to be a prerequisite for acquisition of the totipotency exclusive to these cells (Yamaji et al., 2008). Prdm14 appears to be a key factor in the early steps of germ-cell reprogramming, and shows specific expression in PGCs and pluripotent stem cells. So far, a total of three genes appear key to the unique epigenetic reprogramming and reversion of cells in the epiblast to a totipotent state: blimp1, prdm14, and the transcriptional regulator ap2γ establish an essential chromatin landscape leading to PGC specification (Magnúsdóttir et al., 2012). Following induction of blimp1, prdm14, and ap2γ in the presumptive germ cells, several pluripotency/reprogramming factors are specifically induced in the cells, including klf2 (Kruppel-like factor 2), sox2, and nanog (Kurimoto et al., 2008).
More recently, the Surani group has taken a cell-culture approach to dissect the mechanism of epigenetic regulation of epiblast cells in the mouse embryo and their capacity for PGC development (Gillich et al., 2012). Mouse genetics has provided crucial insights into the mechanisms of inductive PGC specification, yet the epigenetic mechanisms used by germ cells are poorly understood. Moreover, working on germ cells in mice has inherent challenges, especially with the limited numbers of PGCs and the lack of access to the embryo following implantation. Maintaining mouse PGCs in vitro is currently not possible, thus studies manipulating stem cell cultures have become prominent surrogates in studies of PGC development in lieu of the in vivo PGCs. While this comes with complications in interpretations, as does any in vitro model of an in vivo process, it allows additional approaches and more rapid experimentation than otherwise possible. Assays using epiblast stem cells (EpiSCs) are now being used to investigate the role of germline factors in epigenetic reprogramming (Gillich et al., 2012). EpiSCs self-renew continuously, can be expanded to large quantities, and are easy to manipulate. Further, EpiSCs can transition (revert) to ESC-like cells and at that point can undergo specification to unipotent PGCs upon exposure to BMP4 (Hayashi and Surani, 2009). Thus, the reversion of EpiSCs to ESCs represents a more tractable system for testing the effects of germline factors on epigenetic reprogramming. This system was used recently to identify a synergistic effect of Prdm14 and Klf2 in EpiSC reversion that includes rapid and efficient X-chromosome reactivation and loss of DNA methylation (Gillich et al., 2012). Notable here is that the factors work together for this transition, and are individually ineffective in this endpoint. Klf2 is a target of the Prdm14 complex that is induced in the PGC genome when epigenetic reprogramming leads to a potential for early germ-cell specification. Thus, combinatorial-based epigenetic changes in the small set of cells responding to BMP4 appears to tip a balance of somatic/germline fate. The germline fate enables other pluripotency genes to become active and to contribute in different ways to the commitment of these few cells to the immortal germline lineage.
FURTHER LESSONS FOR GERMLINE DETERMINATION
Induction-based germline specification is widespread through phylogeny, with many of the major phyla having representatives of both inductive and inherited mechanisms of germline specification (Extavour and Akam, 2003). How inclusive is the mouse model of inductive germline specification, particularly in explaining this mechanism as a general phenomenon? Are these same molecular players part of a core gene set required for germline determination in all animals, even in those using inherited mechanisms? Highly conserved orthologs of the blimp1 gene can be found across species showing both inherited and inductive specification (John and Garrett-Sinha, 2008), and stella is restricted to the germline of the placental mammals tested (Cañón et al., 2006). Genes such as vasa, nanos, and piwi also function in mouse PGC fate, but they are expressed and appear to function much later in development. Interestingly, capsuleen/dart5, a homologue of Drosophila prmt5, has been shown to regulate germ-cell specification and spermatocyte specification in females and males, respectively (Gonsalvez et al., 2006; Anne et al., 2007). Dart5 mutants show disrupted localization of tudor, an essential gene for polar granule assembly and pole cell specialization in Drosophila (Thomson and Lasko, 2004). This work, along with studies in C. elegans (Barbee and Evans, 2006), provides evidence that Sm proteins, core components of small nuclear ribonucleoproteins (smRNPs) such as dart5, are required for germ-cell specification and maintenance. It is important to note that the above holds true even for organisms undergoing inherited specification via transcriptional and post-transcriptional silencing. Blimp1 also acts to regulate chordin expression in the gastrula organizer and cell-fate specification in Danio, another organism that uses inherited determination (Wilm and Solnica-Krezel, 2005; Tada et al., 2012). Furthermore, the discovery of lin28 as a regulator of PGC formation in mice identified a gene that plays a role across phyla—from regulation of developmental timing in C. elegans (Moss et al., 1997) to germ cells in the human fetal ovary (Childs et al., 2012; see Moss and Tang, 2003 for the conservation of lin28). A link between lin28 and blimp1/prdm14 in other species would further reveal the latter’s role as a master regulator of germ-cell development.
The one factor consistent across blimp1 homologues is that they all act as transcriptional repressors of other genes. It would make sense that such a gene would be required to regulate germ-cell development in an organism where, for the most part, repression of somatic genes is essential for a germline fate. In inherited determination, however, the set of signals for PGC formation is already established in the germ plasm. Presumably, it works in an organism with a more labile nucleus in its developmental program, and is less dependent on repression of instructions that would otherwise lead cells down a somatic path. Drosophila blimp1 (dblimp1) has been shown to play a role in developmental timing (Agawa et al., 2007), but not in germ-cell formation. It seems that the autonomous mode of determination obviates a need for blimp1/prdm14 as a regulator of PGC formation. Thus, the role of blimp1 and prdm14 as transcriptional repressors in the mouse germline helps to distinguish mechanism between the two processes of germline determination, of an inherited process into a cell with a naïve nucleus versus an inductive process in which the nucleus has a history of gene expression, much of which needs to be repressed.
So the question remains: how conserved is the molecular program for animals using induction-based germline specification? Further, what is required to transition from an inductive to an inherited program? This transition appears to have occurred many times in evolution (Extavour and Akam, 2003), so in principle, it must be minimal. Some models of germline determination suggest that this ancient mechanism was replaced through several independent events by a mechanism relying largely on the same germline determinants being localized, and thereby directing fate determination on those cells that acquire them (Extavour, 2007). Another hypothesis is that when gene regulation networks (GRNs) are invoked early as in maternally localized determinants for germline determination, the embryo is more able to change and create new morphologies in the organism (Johnson et al., 2003, 2011; Crother et al., 2007). One selection mechanism for inherited determination may be a relatively earlier germline acquisition, perhaps enabling faster development and therefore an earlier impact on competition in the reproductive niche.
Interestingly, a molecular explanation for a switch between the two mechanisms has been found in holometabolous (“higher”) insects. The evolution of oskar correlates with a switch in germline determination from an inductive to an inherited mechanism (Lynch et al., 2011; reviewed in Extavour, 2011). Remarkably, upon acquisition of oskar (and possibly other characters in the animal) and conversion from a basal inductive-based mechanism to an inheritance-based mechanism, some species subsequently lose functionality of oskar and revert back to an inductive-based mechanism (Lynch et al., 2011). An important goal over the next several years will be to learn how diverse animals using inductive specification accomplish this task. With that molecular information in hand, hopefully investigators will be able to test for consensus mechanisms, if they even exist. From there, one might be able to compare the canonical inheritance next to the canonical inductive programs and see how evolution has accomplished this essential feat so many times, in so many different animals.
It is important, however, to keep in mind that the various processes that make up an inheritance- or inductive-based pathway of germline determination are each likely variable in their roles for each organism. These characteristics are also probably a continuum of differences between organisms (Fig. 2). Evolutionary changes may thus individually work on one process or another, and only once a threshold is exceeded in the overall sum of the individual mechanisms does the embryo become categorized by an investigator as an inheritance- or inductive-based organism. Sea urchins are thought of classically as an inductive-based organism, but also help us understand the continuum model of germ-line determination. Manipulations of the early sea urchin embryo by Sven Horstadius in the early 1900s defined many of the concepts of signaling centers, embryonic signaling gradients, and cell-fate determination based on cell interactions (see Horstadius, 1973). Yet germ-cell determination in this animal appears to have many features of inheritance (autonomy) even though no localized germ plasm or germinal granules are found in the egg or early embryo (Juliano et al., 2006). For example, when the precursor cells (micromeres), of what appears to be the PGCs (small micromere derivatives), are placed in culture, the “germ-cell features” of the PGCs (specific vasa and nanos expression) are still acquired and are on schedule with the germ cells in the embryo (Yajima and Wessel, 2012). Separating early blastomeres and culturing them individually leads to small but complete embryos, including the fractional number of germ cells normally formed by the blastomere. Following the third cell division, the germ-cell precursors form only in the vegetal-derived blastomeres, suggesting that localized germ-cell determinants are present in the vegetal pole (Voronina et al., 2008). Further, if the small micromeres (PGCs) are removed from the early embryo, the animal still develops perfectly well, metamorphoses from a normal looking larva, and develops gonads, but does not show any indication of germ-cell development (Yajima and Wessel, 2011). This is analogous to the phenomenon in Drosophila in which pole cells are removed genetically (Kobayashi et al., 1996). Yet when the micromeres (PGC precursor cells) of the sea urchin embryos are removed, the embryos compensate with a marked up-regulation of vasa throughout the embryo and a gradual restriction of Vasa to endodermal regions, and metamorphosis into gravid adults (Ransick et al., 1996). Thus, the embryos have retained some features of compensatory germline determination, something analogous to that in mice and suggestive of an original inheritance-based mechanism. In sea stars, close relatives of the sea urchin, the putative germline (derivatives of the posterior enterocoel) does not appear until much later in development. The embryos have uniform vasa expression until well after gastrulation, and then it gradually becomes restricted to the endodermal region, and finally enriched in cells of the posterior enterocoel. The above molecular behavior is reminiscent of the sea urchin embryo upon removal of their micromeres (Voronina et al., 2008), and may reflect the retention of characters for inductive germline fate “on the way” to an inherited mechanism.
The compensatory germline activity in the sea urchin is not unique. The parasitic wasp Pimpla turionellae, for example, has distinct germinal granules (oosomes) that are acquired by the early-forming pole cells that give rise to the germline (Bronskill, 1959). Yet, when the oosomes are removed, the animal re-acquires germ cells by compensatory development (from an unknown lineage) to become fully fertile (Achtelig and Krause, 1971). Thus, a range of features may be present in embryos, some revealed only by experimentation, that suggest a broad sharing of mechanistic pieces between the extremes of inheritance and inductive germline determination. We argue here that a continuum of characters exists within the realm of inheritance- and inductive-based germline determination mechanisms that would enable evolutionary transitions to occur many times in phylogeny. Perhaps once we have a better portrait of the molecular players and their functions in the event of germline determination, we will be able to approach the more overarching questions of evolutionary transitions for this important process in the life cycle of an organism.
A strong comparison can be drawn between the process of germline determination and sex determination. Sex determination in vertebrates is the process that directs development of the testes and ovaries from undifferentiated gonads, and the subsequent secondary sexual characteristics of the organism result from sexually dimorphic hormonal control of development (Gilbert, 2010). Two mechanisms for sex determination exist in vertebrates: genetic/genotypic sex determination, in which the sex of the organisms is determined by which chromosomes the zygote acquires, and environmental sex determination, in which external conditions initially direct gonad development into male or female outcomes (Gilbert, 2010). With the exception of the mole vole (Soullier et al., 1998) and spiny rat (Just et al., 1995), all mammals use the Sry gene for male sex determination. This gene is a member of the high mobility group (HMG) DNA binding factor family that is necessary and sufficient for testis development (Sinclair et al., 1990; Koopman et al., 1991). sry leads to expression of sox9 (another member of the HMG-class of DNA-binding proteins; Graves, 1998), and a positive feedback loop enhances the process, including members of the fibroblast growth factor (fgf 9) that enhances sox9 expression (and which down-regulates sry), up regulates anti-Müllerian hormone (Amh), and down-regulates the wnt/beta-catenin pathway.
Development of an ovary in mammals instead involves expression of respondin-1 and wnt1, which enhance the beta-catenin pathway and, in concert with foxl2, repress the sox9/fgf9 amplifying loop. In many non-mammalian vertebrates, sex determination is also chromosomally based and uses many of the same pathway components to generate ovaries and testis, but the process involves initiation of a cascade in a system evolved from the dmrt1 gene (Matsuda et al., 2002, 2007; Yoshimoto et al., 2008; Smith et al., 2009; Mawaribuchi et al., 2012).
Reptiles instead use temperature (one form of environmental sex determination) to initiate development of the gonads, leading to subsequent sex hormonal dimorphisms (Nelson et al., 2004; Sarre et al., 2004; Crews, 2009). Although this environment-based mechanism of sex determination is less well understood than gene-based mechanisms, it is functional in most reptiles and relies on a narrow temperature (a few degrees centigrade) over a short window of time (less than 10% of the developmental period) to direct development of one or another sex in the offspring (Janzen and Paukstis, 1991).
Remarkably, although many reptiles use this external cue for stimulating divergence between male and female development, they use many of the same genes involved in the pathway of sex determination as do the sry or dmrt1 genetic sex determination mechanisms. These include sox9, sf1, dax1, amh, and wt-1, which are each expressed in gonads during their genesis in a variety of reptilian species (Wibbels et al., 1991; Crews et al., 1989, 1991; Spotila et al., 1998; Smith et al., 1999; Kettlewell et al., 2000; Western and Sinclair, 2001).
Genetic and temperature-dependent sex determination appears to function along a continuum of states in animals demonstrated well in reptiles (Sarre et al., 2004; Angelopoulou et al., 2012). In that taxon, sex in some species is determined primarily by genetic mechanisms, some by a combination of genetic and temperature mechanisms, and others primarily by temperature. It is believed that genetic and temperature-dependent sex determination mechanisms have evolved independently on several occasions, leading to this continuum of diversity in initiation of a common pathway. This is true also of the germline determination pathway: a core set of genes (including vasa, nanos, and piwi) are essential elements in the germline pathway used by all animals, yet their utilization differs greatly and is activated differently depending on the animal. These different extremes include the inheritance- and induction-based mechanisms and also follow a continuum of features that enable change over time from the reliance on one mechanism to another many independent times during evolution. A better understanding of the reproductive niche, not just the environmental niche of the organism, will be important for understanding the origins and transitions of these mechanisms.
ACKNOWLEDGMENTS
The authors thank Dr. Celina Juliano for helpful comments in the construction of this work, Rachel Whitaker of the Brown University Writing Center for inputs on the manuscript, and the two expert reviewers of this manuscript who stimulated the sculpting of several new concepts for us. We gratefully acknowledge support for this work from the National Institutes of Health (HD028152) and the National Science Foundation (IOS-1120972) grants to GMW.
Abbreviations
- blimp
B-lymphocyte induced maturation protein
- bmp
bone morphogenetic protein
- dpc
days post-coitum
- ESCs
embryonic stem cells
- prdm
positive regulatory domain l-binding factor
- PGC
primordial germ cell
- prmt
protein arginine methyltransferase
- TNAP
tissue non-specific alkaline phosphatase
Footnotes
Not all organisms have germline stem cells, and not all animals reproduce sexually.
The field of germline determination has many terms referring to these same concepts. This often happens when many investigators study diverse organisms over lengthy, historical times. In terms of the two extreme germline determination mechanisms, the broad concept of the inherited mechanisms may also be referred to as localized, autonomous, and pre-determined; the mechanism in the opposite extreme is sometimes referred to as epigenesis, conditional, non-autonomous, or dependent. Here we use the terms “inherited” and “inductive,” respectively. We find that these terms are more effective in communicating the broad diversity of germline determination mechanisms and amply contrast the processes used without restricting the definitions. For example, presumptive germline cells are seldom cultured in isolation to test autonomy, and in only a few cases have experiments been conducted to test the conditional nature of the cell fate of each cell that may or may not become a PGC. Often, simply the lack of localized candidate gene products is used to conclude that the embryo of a particular species uses an inductive cell fate determination mechanism. Furthermore, the term “inherited” emphasizes the maternal contribution to the germline, implying an early determination mechanism, and is distinct from the opposite extreme in which germline cell fate determination occurs later in development and results from intercellular communication.
REFERENCES
- Achtelig M, Krause G. Experiments on uncleared egg of Pimpla turionellae L. (Hymenoptera) for functional analysis of oosome region. Wilhelm Roux Arch Entwickl Mech Org. 1971;167:164–182. doi: 10.1007/BF00577038. [DOI] [PubMed] [Google Scholar]
- Agawa Y, Sarhan M, Kageyama Y, Akagi K, Takai M, Hashiyama K, Wada T, Handa H, Iwamatsu A, Hirose S, Ueda H. Drosophila Blimp-1 is a transient transcriptional repressor that controls timing of the ecdysone-induced developmental pathway. Mol Cell Biol. 2007;27:8739–8747. doi: 10.1128/MCB.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS ONE. 2010;5:e10531. doi: 10.1371/journal.pone.0010531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Anderson R, Copeland TK, Scholer H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mech Dev. 2000;91:61–68. doi: 10.1016/s0925-4773(99)00271-3. [DOI] [PubMed] [Google Scholar]
- Angelopoulou R, Lavranos G, Manolakou P. Sex determination strategies in 2012: Towards a common regulatory model? Reprod Biol Endocrinol. 2012;10:13. doi: 10.1186/1477-7827-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyl-transferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Arnold S, Robertson EJ. Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germ line development in Drosophila embryos. Nat Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–578. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñá J. Planarian neoblasts. Nature. 1981;290:14–15. [Google Scholar]
- Bao S, Leitch HG, Gillich A, Nichols J, Tang F, Kim S, Lee C, Zwaka T, Li X, Surani MA. The germ cell determinant blimp1 is not required for derivation of pluripotent stem cells. Cell Stem Cell. 2012;11:110–117. doi: 10.1016/j.stem.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Evans TC. The Sm proteins regulate germ cell specification during early C. elegans embryogenesis. Dev Biol. 2006;291:132–143. doi: 10.1016/j.ydbio.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Berger F, Twell D. Germline specification and function in plants. Annu Rev Plant Biol. 2011;62:461–484. doi: 10.1146/annurev-arplant-042110-103824. [DOI] [PubMed] [Google Scholar]
- Bikoff EK, Robertson EJ. One PRDM is not enough for germ cell development. Nat Genet. 2008;40:934–935. doi: 10.1038/ng0808-934. [DOI] [PubMed] [Google Scholar]
- Bikoff EK, Morgan MA, Robertson EJ. An expanding job description for Blimp-1/PRDM1. Curr Opin Genet Dev. 2009;19:379–385. doi: 10.1016/j.gde.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Billett FS, Adam E. The structure of the mitochondrial cloud of Xenopus laevis oocytes. J Embryol External Morphol. 1976;36:697–710. [PubMed] [Google Scholar]
- Birkhead TR. Promiscuity: An evolutionary history of sperm competition. Cambridge, MA: Harvard UP; 2000. [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, Dosch R. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol. 2009;19:414–422. doi: 10.1016/j.cub.2009.01.038. [DOI] [PubMed] [Google Scholar]
- Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Goodheart M, Liao M, Page DC. Dppa3/Pgc7/stella is a maternal factor and is not required for germ cell specification in mice. BMC Dev Biol. 2004;4:2. doi: 10.1186/1471-213X-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braat AK, van de Water S, Goos H, Bogerd J, Zivkovic D. Vasa protein expression and localization in the zebrafish. Mech Dev. 2000;95:271–274. doi: 10.1016/s0925-4773(00)00344-0. [DOI] [PubMed] [Google Scholar]
- Braat AK, van de Water S, Korving J, Zivkovic D. A zebrafish vasa morphant abolishes vasa protein but does not affect the establishment of the germline. Genesis. 2001;30:183–185. doi: 10.1002/gene.1060. [DOI] [PubMed] [Google Scholar]
- Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. Prmt5 (janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem. 2001;276:32971–32976. doi: 10.1074/jbc.M105412200. [DOI] [PubMed] [Google Scholar]
- Breitwieser W, Markussen FH, Horstmann H, Ephrussi A. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 1996;10:2179–2188. doi: 10.1101/gad.10.17.2179. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Bronskill JF. Embryology of Pimpla turnionellae L. (Hymenoptera: Ichneumonidae) Can J Zool. 1959;37:655–688. [Google Scholar]
- Buss L. The evolution of individuality. Princeton, NJ: Princeton University Press; 1987. [Google Scholar]
- Buss DM. The evolution of desire: Strategies of human mating. New York, NY: Basic Books; 1994. [Google Scholar]
- Bykova O, Chuine I, Morin X, Higgins SI. Temperature dependence of the reproduction niche and its relevance for plant species distributions. J Biogeogr. 2012;39:2191–2200. [Google Scholar]
- Canon S, Herranz C, Manzanares M. Germ cell restricted expression of chick Nanog. Dev Dyn. 2006;235:2889–2894. doi: 10.1002/dvdy.20927. [DOI] [PubMed] [Google Scholar]
- Chang DH, Calame KL. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (BLIMP-1) during mouse embryonic development. Mech Dev. 2002;117:305–309. doi: 10.1016/s0925-4773(02)00189-2. [DOI] [PubMed] [Google Scholar]
- Childs AJ, Kinnell HL, He J, Anderson RA. LIN28 is selectively expressed by primordial and pre-meiotic germ cells in the human fetal ovary. Stem Cells Dev. 2012;21:2343–2349. doi: 10.1089/scd.2011.0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquoine AD. The identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat Rec. 1954;118:135–146. doi: 10.1002/ar.1091180202. [DOI] [PubMed] [Google Scholar]
- Coward SJ. Chromatoid bodies in somatic cells of the planarian: Observations on their behavior during mitosis. Anat Rec. 1974;180:533–545. doi: 10.1002/ar.1091800312. [DOI] [PubMed] [Google Scholar]
- Crews D. Mode and tempo in environmental sex determination in vertebrates. Semin Cell Dev Biol. 2009;20:251–255. doi: 10.1016/j.semcdb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Crews D, Wibbels T, Gutzke WHN. Action of sex steroid hormones on temperature-induced sex determination in the snapping turtle (Chelydra serpentina) Gen Comp Endocrinol. 1989;75:159–166. doi: 10.1016/0016-6480(89)90042-7. [DOI] [PubMed] [Google Scholar]
- Crews D, Bull JJ, Wibbels T. Estrogen and sex reversal in turtles: Dosages producing both sexes produce few intersexes. Gen Comp Endocrinol. 1991;81:357–364. doi: 10.1016/0016-6480(91)90162-y. [DOI] [PubMed] [Google Scholar]
- Crother BI, White ME, Johnson AD. Inferring developmental constraint and constraint release: Primordial germ cell determination mechanisms as examples. J Theor Biol. 2007;248:322–330. doi: 10.1016/j.jtbi.2007.05.035. [DOI] [PubMed] [Google Scholar]
- Draper BW, McCallum BM, Moens CB. Nanos1 is required to maintain oocyte production in adult zebrafish. Dev Biol. 2007;305:589–598. doi: 10.1016/j.ydbio.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcova-Hills G, Capel B. Chapter 6: Development of germ cells in the mouse. Curr Top Dev Biol. 2008;83:185–212. doi: 10.1016/S0070-2153(08)00406-7. [DOI] [PubMed] [Google Scholar]
- Durcova-Hills G, Tang F, Doody G, Tooze R, Surani MA. Reprogramming primordial germ cells into pluripotent stem cells. PLoS ONE. 2008;3:e3531. doi: 10.1371/journal.pone.0003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R. Induction of germ-cell formation by Oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, Marín de Evsikova C. Evolutionary origin and phylogenetic analysis of the novel oocyte-specific eukaryotic translation initiation factor 4E in Tetrapoda. Dev Genes Evol. 2009;219:111–118. doi: 10.1007/s00427-008-0268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B, Schwager EE, Extavour CG. The molecular machinery of germ line specification. Mol Reprod Dev. 2010;77:3–18. doi: 10.1002/mrd.21091. [DOI] [PubMed] [Google Scholar]
- Extavour CG. Evolution of the bilaterian germ line: Lineage origin and modulation of specific organisms. Integr Comp Biol. 2007;47:770–785. doi: 10.1093/icb/icm027. [DOI] [PubMed] [Google Scholar]
- Extavour CG. Long-lost relative claims orphan gene: Oskar in a wasp. PLoS Genet. 2011;7:e1002045. doi: 10.1371/journal.pgen.1002045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: Epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Lynch M. Non-adaptive origins of interactome complexity. Nature. 2011;474:502–506. doi: 10.1038/nature09992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Forsthoefel DJ, James NP, Escobar DJ, Stary JM, Vieira AP, Waters FA, Newmark PA. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev Cell. 2012;23:691–704. doi: 10.1016/j.devcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Wyce A, Paushkin S, Abel L, Rappsilber J, Mann M, Dreyfuss G. A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem. 2011;277:8243–8247. doi: 10.1074/jbc.M109984200. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Wang JT, Motegi F, Seydoux G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 2010;330:1685–1689. doi: 10.1126/science.1193697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Arkov A. Next generation organelles: Structure and role of germ granules in germline. Mol Reprod Dev. 2012 doi: 10.1002/mrd.22115. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S. Rapid evolution of reproductive barriers driven by sexual conflict. Nature. 2000;403:886–889. doi: 10.1038/35002564. [DOI] [PubMed] [Google Scholar]
- Gilbert S. Developmental biology. 9th edition. MA: Sinauer Pub; 2010. [Google Scholar]
- Gillich A, Hayashi K. Switching stem cell state through programmed germ cell reprogramming. Development. 2011;81:281–291. doi: 10.1016/j.diff.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Gillich A, Bao S, Grabole N, Hayashi K, Trotter MW, Pasque V, Magnúsdóttir E, Surani MA. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell. 2012;10:425–439. doi: 10.1016/j.stem.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol. 2006;16:1077–1089. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: A critique of the adaptationist programme. Proc R Soc London. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- Graham PL, Kimble J. The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics. 1993;133:919–931. doi: 10.1093/genetics/133.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. Interactions between SRY and SOX genes in mammalian sex determination. BioEssays. 1998;20:264–269. doi: 10.1002/(SICI)1521-1878(199803)20:3<264::AID-BIES10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. Aslicer-mediated mechanism for repeat-associated siRNA 50 end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Gupta YK, Lee TH, Edwards TA, Escalante CR, Kadyrova LY, Wharton RP, Aggarwal AK. Co-occupancy of two Pumilio molecules on a single hunchback NRE. RNA. 2009;15:1029–1035. doi: 10.1261/rna.1327609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Wessel GM. Vasa genes: Emerging roles in the germ line and in multipotent cells. BioEssays. 2010;32:626–637. doi: 10.1002/bies.201000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Yajima M, Juliano C, Wessel GM. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryo-genesis. Dev Biol. 2011;349:440–450. doi: 10.1016/j.ydbio.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Sudo T, Mikami Y, Otani M, Takano M, Tsuda H, Itamochi H, Katabuchi H, Ito M, Nishimura R. Germ cell specific protein VASA is over-expressed in epithelial ovarian cancer and disrupts DNA damage-induced G2 checkpoint. Gynecol Oncol. 2008;111:312–319. doi: 10.1016/j.ygyno.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55:577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Surani MA. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development. 2009;136:3549–3556. doi: 10.1242/dev.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, de Sousa Lopes SM, Surani MA. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- Hernández G, Han H, Gandin V, Fabian L, Ferreira T, Zuberek J, Sonenberg N, Brill JA, Lasko P. Eukaryotic initiation factor 4E-3 is essential for meiotic chromosome segregation, cytokinesis and male fertility in Drosophila. Development. 2012;139:3211–3220. doi: 10.1242/dev.073122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird SN, Paulsen JE, Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: Cell-type-specific mechanisms for cytoplasmic localization. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- Hori I. An ultrastructural study of the chromatoid body in planarian regenerative cells. J Electron Microsc. 1982;31:63–72. [Google Scholar]
- Horstadius S. Experimental embryology of echinoderms. Oxford: Clarendon; 1973. [Google Scholar]
- Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci USA. 1974;71:1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germ line genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- Janzen FJ, Paukstis GL. Environmental sex determination in reptiles: Ecology, evolution, and experimental design. Q Rev Biol. 1991;66:149–179. doi: 10.1086/417143. [DOI] [PubMed] [Google Scholar]
- John SA, Garrett-Sinha LA. blimp1: A conserved transcriptional repressor critical for differentiation of many tissues. Exp Cell Res. 2008;315:1077–1084. doi: 10.1016/j.yexcr.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Drum M, Bachvarova RF, Masi T, White ME, Crother BI. Evolution of predetermined germ cells in vertebrate embryos: Implications for macroevolution. Evol Dev. 2003;5:414–431. doi: 10.1046/j.1525-142x.2003.03048.x. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Richardson E, Bachvarova RF, Crother BI. Evolution of the germ line-soma relationship in vertebrate embryos. Reproduction. 2011;141:291–300. doi: 10.1530/REP-10-0474. [DOI] [PubMed] [Google Scholar]
- Judson O. Dr. Tatiana’s sex advice to all creation: The definitive guide to the evolutionary biology of sex by Olivia Judson. New York: Henry Holt and Company publishers; 2002. [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300:406–415. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Wang J, Lin H. Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just W, Rau W, Vogul W, Akhverdian M, Fredga K, Graves JA, Lyapunova E. Absence of Sry in species of the vole Ellobius. Nat Genet. 1995;11:117–118. doi: 10.1038/ng1095-117. [DOI] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, Wharton RP. Translational control of maternal cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868–879. doi: 10.1101/gad.5.5.868. [DOI] [PubMed] [Google Scholar]
- Kettlewell JR, Raymond CS, Zarkower D. Temperature-dependent expression of turtle Dmrt1 prior to sexual differentiation. Genesis. 2000;26:174–178. [PubMed] [Google Scholar]
- Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149:875–888. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 1996;380:708–711. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanosrelated gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota SK, Feil R. Epigenetic transitions in germ cell development and meiosis. Dev Cell. 2010;19:675–686. doi: 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, Totoki Y, Shibata T, Kimura T, Nakatsuji N, Noce T, Sasaki H, Nakano T. MVH in pi RNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M. Complex genome-wide transcription dynamics orchestrated by blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Singh A, King ML. Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development. 2012;139:1476–1486. doi: 10.1242/dev.079608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange UC, Saitou M, Western PS, Barton SC, Surani MA. The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol. 2003;3:1–11. doi: 10.1186/1471-213X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange UC, Adams DJ, Lee C, Barton SC, Schneider R, Bradley A, Surani MA. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol Cell Biol. 2008;15:4688–4696. doi: 10.1128/MCB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Hage WJ. Clonal analysis on the origin of primordial germ cells in the mouse. Ciba Found Symp. 1994;182:68–84. doi: 10.1002/9780470514573.ch5. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Dunn RR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development (Cambridge, England) 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lin Y, Wong K, Calame KL. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cells differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- Linder P, Lasko P. Bent out of shape: RNA unwinding by the DEAD-Box helicase vasa. Cell. 2006;125:219–221. doi: 10.1016/j.cell.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schnier J, Slonimski PP. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- Liu N, Han H, Lasko P. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev. 2009;23:2742–2752. doi: 10.1101/gad.1820709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007;104:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Lynch JA, Ozuak O, Khila A, Abouheif E, Desplan C, Roth S. The phylogenetic origin of oskar coincided with the origin of maternally provisioned germ plasm and pole cells at the base of the Holometabola. PLoS Genet. 2011;7:e1002029. doi: 10.1371/journal.pgen.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor GR, Zambrowicz BP, Soranio P. Tissue nonspecific alkaline phosphatase is expressed in both embryonic and extraembryonic lineages during mouse embryogenesis but is not required for migration of primordial germ cells. Development. 1995;121:1487–1496. doi: 10.1242/dev.121.5.1487. [DOI] [PubMed] [Google Scholar]
- Magnúsdóttir E, Gillich A, Grabole N, Surani MA. Combinatorial control of cell fate and reprogramming in the mammalian germline. Curr Opin Genet Dev. 2012;22:466–474. doi: 10.1016/j.gde.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. Assembly of the Drosophila germ plasm. Int Rev Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol. 2008;321:40–50. doi: 10.1016/j.ydbio.2008.05.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins G, Calame KL. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, Hori H, Hamaguchi S, Sakaizumi M. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Shinomiya A, Kinoshita M, Suzuki A, Kobayashi T, Paul-Prasanth B, Lau EL, Hamaguchi S, Sakaizumi M, Nagahama Y. DMY gene induces male development in genetically female (XX) medaka fish. Proc Natl Acad Sci USA. 2007;104:3865–3870. doi: 10.1073/pnas.0611707104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM. Lin28 lets BLIMP1 take the right course. Dev Cell. 2009;17:160–161. doi: 10.1016/j.devcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Mawaribuchi S, Yoshimoto S, Ohashi S, Takamatsu N, Ito M. Molecular evolution of vertebrate sex-determining genes. Chromosome Res. 2012;20:139–151. doi: 10.1007/s10577-011-9265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren A, Lawson KA. How is the mouse germ-cell lineage established? Differentiation. 2005;73:435–437. doi: 10.1111/j.1432-0436.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- Medrano JV, Ramathal C, Nguyen HN, Simon C, Reijo Pera RA. Divergent RNA-binding proteins, DAZL and VASA, induce meiotic progression in human germ cells derived in vitro. Stem Cells. 2012;30:441–451. doi: 10.1002/stem.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Torres-Padilla M-E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T. Expression and evolutionary conservation of nanos-related genes in Hydra. Dev Genes Evol. 2000;210:591–602. doi: 10.1007/s004270000105. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germ line expression in Hydra. Dev Genes Evol. 2001;211:299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- Monk M, Harper MI. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature. 1979;281:311–313. doi: 10.1038/281311a0. [DOI] [PubMed] [Google Scholar]
- Morita M. Observation on the fine structure of the neoblast and its cell division in the regenerating planaria. Sci Rep Tohoku Univ ser 4 (Biol) 1967;33:399–406. [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Nagamatsu G, Kosaka T, Kawasumi M, Kinoshita T, Takubo K, Akiyama T, Sudo T, Kobayashi T, Oya M, Suda T. A germ cell-specific gene, Prmt5, works in somatic cell reprogramming. J Biol Chem. 2011;286:10641–10648. doi: 10.1074/jbc.M110.216390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 1996;274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu Y-J, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Orugra A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5 mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Nelson NJ, Cree A, Thompson MB, Keall SN, Daucherty CH. Temperature-dependent sex determination in tuatara. In: Valenzuela N, Lance VA, editors. Temperature-dependent sex determination in vertebrates. Washington: Smithsonian Institution; 2004. pp. 53–58. [Google Scholar]
- Newmark PA, Wang Y, Chong T. Germ cell specification and regeneration in planarians. Cold Spring Harb Symp Quant Biol. 2008;73:573–581. doi: 10.1101/sqb.2008.73.022. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA. blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Sano M, Shigeta M, Yamanaka K, Saitou M. A comprehensive, non-invasive visualization of primordial germ cell development in mice by the Prdm1-mVenus and Dppa3-ECFP double transgenic reporter. Reproduction. 2008;136:503–514. doi: 10.1530/REP-08-0053. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Saitou M, Barton SC, Thresher R, Dixon JPC, Zahn D, Colledge WH, Carlton MBL, Nakano T, Surani MA. Stella is a maternal effect gene required for normal early development in mice. Curr Biol. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Pederson KJ. Cytological studies on the planarian neoblast. Z Zellforsch Mikrosk Anat (Vienna, Austria: 1948) 1959;50:799–817. doi: 10.1007/BF00342367. [DOI] [PubMed] [Google Scholar]
- Pek JW, Kai T. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr Biol. 2011a;21:39–44. doi: 10.1016/j.cub.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Pek JW, Kai T. DEAD-box RNA helicase Belle/DDX3 and the RNA interference pathway promote mitotic chromosome segregation. Proc Natl Acad Sci USA. 2011b;108:12007–12012. doi: 10.1073/pnas.1106245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister D, De Mulder K, Philipp I, Kuales G, Hrouda M, Eichberger P, Borgonie G, Hartenstein V, Ladurner P. The exceptional stem cell system of Macrostomum lignano: Screening for gene expression and studying cell proliferation by hydroxyurea treatment and irradiation. Front Zool. 2007;4:9. doi: 10.1186/1742-9994-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister D, De Mulder K, Hartenstein V, Kuales G, Borgonie G, Marx F, Morris J, Ladurner P. Flatworm stem cells and the germ line: Developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev Biol. 2008;319:146–159. doi: 10.1016/j.ydbio.2008.02.045. [DOI] [PubMed] [Google Scholar]
- Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame KL. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–632. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- Ransick A, Cameron RA, Davidson EH. Postembryonic segregation of the germ line in sea urchins in relation to indirect development. Proc Natl Acad Sci USA. 1996;93:6759–6763. doi: 10.1073/pnas.93.13.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E. The function and regulation of vasa-like genes in germ cell development. Genome Biol. 2000;1:1–6. doi: 10.1186/gb-2000-1-3-reviews1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Chee KJ, Kim TH, Maniatis T. PRD1-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SM. Evolving responsively: Adaptive mutation. Nat Rev Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- Saitou M. Germ cell specification in mice. Curr Opin Genet Dev. 2009;19:386–395. doi: 10.1016/j.gde.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Saitou M, Yamaji M. Germ cell specification in mice: Signaling, transcription regulation, and epigenetic consequences. Reproduction. 2010;139:931–942. doi: 10.1530/REP-10-0043. [DOI] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Sarre SD, Georges A, Quinn A. The ends of a continuum: Genetic and temperature-dependent sex determination in reptiles. BioEssays. 2004;26:639–645. doi: 10.1002/bies.20050. [DOI] [PubMed] [Google Scholar]
- Sato M, Kimura T, Kurokawa K, Fujita Y, Abe K, Masuhara M, Yasunaga T, Ryo A, Yamamoto M, Nakano T. Identification of PGC7, a new gene expressed specifically in preimplantation embryos and germ cells. Mech Dev. 2002;113:91–94. doi: 10.1016/s0925-4773(02)00002-3. [DOI] [PubMed] [Google Scholar]
- Sato K, Shibata N, Orii H, Amikura R, Sakurai T, Agata K, Kobayashi S, Watanabe K. Identification and origin of the germline stem cells as revealed by the expression of nanos-related gene in planarians. Dev Growth Differ. 2006;48:615–628. doi: 10.1111/j.1440-169X.2006.00897.x. [DOI] [PubMed] [Google Scholar]
- Sato K, Hayashi Y, Ninomiya Y, Shigenobu S, Arita K, Mukai M, Kobayashi S. Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc Natl Acad Sci USA. 2007;104:7455–7460. doi: 10.1073/pnas.0610052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: Lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K. Expression of vasa (vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol. 1999;206:73–87. doi: 10.1006/dbio.1998.9130. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Smith JL, Wilson JE, Macdonald PM. Overexpression of oskar directs ectopic activation of nanos and presumptive pole cell formation in Drosophila embryos. Cell. 1992;70:849–859. doi: 10.1016/0092-8674(92)90318-7. [DOI] [PubMed] [Google Scholar]
- Smith CA, McClive PJ, Western PS, Reed KJ, Sinclair AH. Evolution—Conservation of a sex-determining gene. Nature. 1999;402:601–602. doi: 10.1038/45130. [DOI] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullier S, Hanni C, Catzeflis F, Berta P, Laudet V. Male sex determination in the spiny rat Tokudaia osimensis (Rodentia: Muridae) is not Sry dependent. Mamm Genome. 1998;9:590–592. doi: 10.1007/s003359900823. [DOI] [PubMed] [Google Scholar]
- Spotila LD, Spotila JR, Hall SE. Sequence and expression analysis of Wt1 and Sox9 in the red-eared slider turtle, Trachemys scripta. J Exp Zool. 1998;281:417–427. doi: 10.1002/(sici)1097-010x(19980801)281:5<417::aid-jez7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Steele RE. The Hydra genome: Insights, puzzles and opportunities for developmental biologists. Int J Dev Biol. 2012;56:535–542. doi: 10.1387/ijdb.113462rs. [DOI] [PubMed] [Google Scholar]
- Stower H. Small RNAs: piRNA surveillance in the C. elegans germline. Nat Rev Genet. 2012;13:518–519. doi: 10.1038/nrg3289. [DOI] [PubMed] [Google Scholar]
- Strome S, Martin P, Schierenberg E, Paulsen J. Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development. 1995;121:2961–2972. doi: 10.1242/dev.121.9.2961. [DOI] [PubMed] [Google Scholar]
- Sun F, Palmer K, Handel MA. Mutation of Eif4g3, encoding a eukaryotic translation initiation factor, causes male infertility and meiotic arrest of mouse spermatocytes. Development. 2010;137:1699–1707. doi: 10.1242/dev.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Hajkova P. Epigenetic reprogramming of mouse germ cells toward totipotency. Cold Spring Harb Symp Quant Biol. 2010;75:211–218. doi: 10.1101/sqb.2010.75.010. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci USA. 2010;107:3594–3599. doi: 10.1073/pnas.0908664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Saba R, Miyoshi K, Morita Y, Saga Y. Interaction between NANOS2 and the CCR4-NOT deadenylation complex is essential for male germ cell development in mouse. PLoS ONE. 2012;7:e33558. doi: 10.1371/journal.pone.0033558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Tada H, Mochii M, Orii H, Watanabe K. Ectopic formation of primordial germ cells by transplantation of the germ plasm direct evidence for germ cell determinant in Xenopus. Dev Biol. 2012;371:86–93. doi: 10.1016/j.ydbio.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takamura K, Fujimura M, Yamaguchi Y. Primordial germ cells originate from the endodermal strand cells in the ascidian Ciona intestinalis. Dev Genes Evol. 2002;212:11–18. doi: 10.1007/s00427-001-0204-1. [DOI] [PubMed] [Google Scholar]
- Tarn PPL, Loebel DA. Gene function in mouse embryo-genesis: Get set for gastrulation. Nat Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tarn PPL, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Matsui Y. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech Dev. 2002;119:S261–S267. doi: 10.1016/s0925-4773(03)00126-6. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H, Tarn PPL. IFITM/Mil/Fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev Cell. 2005;9:745–756. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Tee W-W, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, Surani MA. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24:2772–2777. doi: 10.1101/gad.606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Tsuneyoshi N, Sumi T, Onda H, Nojima H, Nakatsuji N, Suemori H. PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem Biophys Res Commun. 2008;367:899–905. doi: 10.1016/j.bbrc.2007.12.189. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Voronina E. The diverse functions of germline P granules in C. elegans. Mol Reprod Dev. 2012 doi: 10.1002/mrd.22136. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour CG, George S, Oliveri P, McClay D, Wessel GM. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314:276–286. doi: 10.1016/j.ydbio.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zayas RM, Guo T, Newmark PA. Nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci USA. 2007;104:5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park I, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western PS, Sinclair AH. Sex, genes, and heat: Triggers of diversity. J Exp Zool. 2001;290:624–631. doi: 10.1002/jez.1113. [DOI] [PubMed] [Google Scholar]
- Wibbels T, Bull JJ, Crews D. Chronology and morphology of temperature-dependent sex determination. J Exp Zool. 1991;260:371–381. doi: 10.1002/jez.1402600311. [DOI] [PubMed] [Google Scholar]
- Wilczek C, Chitta R, Woo E, Shabanowitz J, Chait BT, Hunt DF, Shechter D. Protein arginine methyltransferase Prmt5-Mep50 methylates histones H2A and H4 and the histone chaperone nucleoplasms in Xenopus laevis eggs. J Biol Chem. 2011;9:42221–42231. doi: 10.1074/jbc.M111.303677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm TP, Solnica-Krezel L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development. 2005;132:393–404. doi: 10.1242/dev.01572. [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Wu X, Ruvkun G. Germ cell genes and cancer. Science. 2010;330:1761–1762. doi: 10.1126/science.1200772. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod. 2006;75:705–716. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. The DEAD-box RNA helicase Vasa functions in embryonic mitotic progression in the sea urchin. Development. 2011;138:2217–2222. doi: 10.1242/dev.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Autonomy in specification of primordial germ cells and their passive translocation in the sea urchin. Development. 2012;139:3786–3794. doi: 10.1242/dev.082230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, Nishida-Umehara C, Matsuda Y, Takamatsu N, Shiba T, Ito M. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]