Abstract
Background
Membrane receptors are frequent targets of cancer therapeutic and imaging agents. However, promising in vitro results often do not translate to in vivo clinical applications. To better understand this obstacle, we measured the expression differences in receptor signatures among several human prostate cancer cell lines and xenografts as a function of tumorigenicity.
Methods
Messenger RNA and protein expression levels for integrin ανβ3, neurotensin receptor 1 (NTSR1), prostate specific membrane antigen (PSMA), and prostate stem cell antigen (PSCA) were measured in LNCaP, C4-2, and PC-3 human prostate cancer cell lines and in murine xenografts using quantitative reverse transcriptase polymerase chain reaction, flow cytometry, and immunohistochemistry.
Results
Stable expression patterns were observed for integrin αν and PSMA in all cells and corresponding xenografts. Integrin β3 mRNA expression was greatly reduced in C4-2 xenografts and greatly elevated in PC-3 xenografts compared with the corresponding cultured cells. NTSR1 mRNA expression was greatly elevated in LNCaP and PC-3 xenografts. PSCA mRNA expression was elevated in C4-2 xenografts when compared with C4-2 cells cultured in vitro. Furthermore, at the protein level, PSCA was re-expressed in all xenografts compared with cells in culture.
Conclusions
The regulation of mRNA and protein expression of the cell-surface target proteins ανβ3, NTSR1, PSMA, and PSCA, in prostate cancer cells with different tumorigenic potential, was influenced by factors of the microenvironment, differing between cell cultures and murine xenotransplants. Integrin ανβ3, NTRS1 and PSCA mRNA expression increased with tumorigenic potential, but mRNA expression levels for these proteins do not translate directly to equivalent expression levels of membrane bound protein.
Keywords: ανβ3, NTSR1, PSCA, PSMA, tumorigenic potential, cells versus xenografts, membrane proteins
Introduction
Prostate cancer is the second most common cancer and the second leading cause of cancer death in men in the United States (1). Prominent and unresolved problems with the clinical management of prostate cancer include the lack of highly specific detection methods and efficient therapeutic interventions. Serum prostate specific antigen (PSA) measurements have been used as a measure of the presence of disease, yet abnormal PSA levels can also result from benign prostatic hyperplasia (BPH), and other non-malignant processes, indicating that PSA measurements lack specificity (2). Biopsies are recommended if abnormal PSA levels are found, but results from the European Randomized Study of Screening for Prostate Cancer imply that there is around a 75% negative biopsy rate using PSA as a diagnostic marker (3,4). These results confirm previous studies that reported a 30-50% false-negative biopsy rate in patients with subsequently confirmed malignancy due to small and inconspicuous lesions (5). Twenty to 40 per cent of prostate cancer patients initially responding to treatment by androgen ablation, prostatectomy or radiation, relapse and ultimately progress to castration resistant disease (6). Subsequent chemotherapeutic options are limited, often inefficient, and prone to side effects due to lack of specificity (7).
The specific targeting of cancer cells has become a unifying theme supporting the development of novel imaging and therapy modes (8). Often, the targeting molecules are antibodies, or peptides, which bind to cell-surface membrane proteins that are specifically-, or over-expressed on malignant cells but not expressed on healthy cells. These innovative targeted therapeutic and diagnostic methods promise to increase both the specificity and efficacy of prostate tumor diagnosis and treatment (9,10) while reducing the side-effects (11,12).
Monofunctional, targeted nanoparticles were developed as magnetic resonance imaging (MRI) and drug delivery agents for detection and therapy of prostate cancer (13). These superparamagnetic iron oxide nanoparticles (SPIONs) and superparamagnetic iron platinum particles (SIPPs), when conjugated to a monoclonal antibody against prostate specific membrane antigen (PSMA), specifically bound to PSMA-positive prostate cancer cells in vitro and generated contrast enhancement in MR images (13). While monofunctional nanoparticles performed well, it was reasonable to expect that the efficacy of imaging and therapeutic agents could be improved by using multiple targeting motifs on a single nanoparticle, because this would markedly increase the affinity of the nanoparticles for their targets. Furthermore, such a multifunctional approach might be required in order to detect and treat advanced tumors that are characterized by increased heterogeneity of target antigen expression (2). Imaging and therapeutic agents simultaneously directed to multiple targets expressed by cancer cells should show increased affinities, effectiveness, and specificities when compared with monofunctional agents. These targeting strategies can be tested in suitable prostate cancer cell models with well-characterized phenotypes, such as the human cell lines LNCaP, C4-2, and PC-3, which feature increasing tumorigenic potential and are widely-used in basic and pre-clinical research (13,14). The androgen dependent LNCaP cells were originally isolated from a lymph node metastasis, but are non-aggressive in in vitro assays and have low tumorigenicity in vivo (13). The C4-2 cells are derivatives of LNCaP cells that were passaged in castrated mice, a procedure rendering them androgen-independent, and more-invasive, characteristics associated with human progressive prostate cancer and moderate tumorigenicity (13). The androgen-independent PC-3 cells were isolated from a bone metastasis in a patient with castration-resistant prostate cancer (CRPC) and consequently display a high tumorigenic potential (13). In order to use these cells for the development of multi-targeted imaging or therapeutic agents, it was important to characterize their membrane antigen expression profiles (membrane receptor signatures) with respect to potential targeting motifs. In the present study, we measured the mRNA and cell-surface protein expression profiles for four membrane bound proteins that are over-expressed in prostate cancer and implicated in cancer progression (2,15-18). These cell-surface proteins included the integrin ανβ3, the neurotensin receptor 1 (NTSR1), PSMA, and prostate stem cell antigen (PSCA) in LNCaP, C4-2, and PC-3 cells. Furthermore, because promising in vitro results with cells often do not translate in vivo to similar results in tumors, we determined the differences in the expression of these receptors between cells cultured in vitro as opposed to cell deposits grown as xenografts in immunocompromised mice in vivo.
This study provides an, as yet unreported, overview of the expression signatures for membrane receptors with targeting potential in prostate cancer cells. Knowledge generated in this study should provide further guidance in assessing the utility of cell lines, animal models, and surface markers for targeting purposes in prostate cancer research. Caution should be exercised when it is assumed that the same cell-surface markers are present on cells and xenografts from these same cells.
Materials and Methods
Materials
The prostate cancer cell lines LNCaP and PC-3 were purchased from the American Tissue Type Collection (Manassas, VA, USA). The C4-2 prostate cancer cell line was a kind gift from Dr. G.N. Thalmann (University of Bern, Switzerland). Anti-PSMA, clone J591 antibody was purchased from Neil H. Bander, MD (Cornell College of Medicine, USA). FITC-labeled mouse IgG1 control antibody was obtained from BD Biosciences (San Jose, CA, USA). FITC-labeled mouse anti-PSMA IgG1, clone 107-1A4 antibody was obtained from Medical & Biological Laboratories Co., Ltd. (Woburn, MA, USA). FITC-labeled mouse anti-PSCA IgG1, clone 7F5 and mouse anti-NTSR1 IgM antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). FITC-labeled mouse anti-CD51/61 IgG1, clone 23C6 and FITC-labeled rat anti-mouse, clone RMM-1 antibodies were obtained from Biolegend (San Diego, CA, USA). Quantum Simply Cellular anti-Mouse IgG-Medium Level epitope-density calibration beads were obtained from Bangs Laboratories, Inc. (Fishers, IN, USA). All other chemicals and supplies were purchased from common manufacturers.
Cell Culture
All cell lines were cultured on 75 mm plastic plates in T-medium with 10% FCS (19) at 37 °C in a humidified, 5% CO2 atmosphere. Upon reaching 90% confluency, cells used for qRT-PCR were detached from the plates with the aid of a 0.5% trypsin solution containing 0.02% EDTA, collected with centrifugation at 700 rpm and stored at -80 °C in PBS. Cells used for flow cytometry were released using 5.0 mM EDTA and pipetted to form a monodisperse suspension. Cells were then washed with PBS, and immediately used.
Xenograft Production
The University of New Mexico Institutional Animal Care and Use Committee approved all experiments involving animals. Three million LNCaP, C4-2, or PC-3 cells in 1:1 (vol/vol) BD Matrigel™ (BD Biosciences, San Jose, CA, USA) were injected into the right flank of 5-8 week old athymic nude male mice (Harlan Sprague Dawley, Frederick, MD, USA). Once the tumors had reached a volume of ∼100 mm3, the mice were euthanized using CO2 asphyxiation. For qRT-PCR, the tumors were excised, flash frozen in liquid nitrogen, and stored at -80 °C. For immunohistochemistry, tumors were excised and fixed in 10% buffered formalin.
Flow Cytometry
The Mean Channel Fluorescence (MCF) of cells stained with fluorescein-isothiocyante-, (FITC)-, labeled IgG1 antibodies was compared with a standard curve generated using the appropriate IgG1 corrected for the IgG1 control fluorescence. The epitope-density calibration beads and 105 cells were stained separately by the addition of 15 μl of the appropriate antibody or control antibody in 100 μl Phosphate Buffered Saline (PBS). Cells and beads were incubated in the dark at room temperature for 15 minutes, washed once with PBS, and resuspended in 200 μl PBS. The calibration beads and cells were analyzed using FL1 on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). IgG1 antibodies against all epitopes were available, except for those against NTSR1, which were only of the IgM class.
Since the epitope-density calibration beads were specific for IgG antibodies, and could not be used for the NTSR1-directed IgM antibodies, we compared the MCF of the cells stained for NTSR1 with that of the secondary antibody alone. One hundred thousand cells were stained by the addition of 15 μl of NTSR1 IgM antibody in 100 μl PBS. Cells were incubated in the dark at room temperature for 15 minutes, washed once with PBS, and resuspended in 100 μl PBS. Fluorescent staining was performed by adding 15 μl of FITC-labeled anti-mouse antibody to the labeled and control, unlabeled cells. These samples were incubated in the dark at room temperature for 15 minutes, washed once with PBS, and resuspended in 200 μl PBS. The calibration beads and cells were analyzed using FL1 on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Messenger RNA (mRNA) Expression Analysis by Quantitative Real Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
Baseline expression of αν, β3, NTSR1, PSMA, and PSCA was determined by qRT-PCR. Cultured cells were recovered as explained above. Tissues (0.1-0.5 mg) were immediately frozen in liquid nitrogen upon resection from the animals and subjected to complete homogenization (15-100 seconds) in isothiocyante containing chaotropic buffer (Qiagen, Valencia, CA, USA) using a rotor-stator homogenizer. Total RNA was extracted using RNEasy kits from Qiagen (Valencia, CA, USA) according to the manufacturer's protocols, and analyzed for concentration and purity using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington DE, USA). The integrity of the 5 S, 18 S, and 28 S ribosomal RNAs was examined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Complementary DNA (cDNA) was prepared using the RETROscript cDNA synthesis kit from Ambion (Austin, TX, USA) using random decamer primers in the presence of an RNase inhibitor (Promega, Madison, WI, USA). TaqMan qRT-PCR was performed using a Roche 480 Light Cycler (Roche, Indianapolis, IN, USA). Primers and probes were designed using sequence information from the National Center for Biotechnology Information (NCBI) and Primer Express Software (Applied Biosystems, Carlsbad, CA, USA). The sequences of the primers and probes are given in Table 1. Five hundred nanograms of cDNA were used per reaction in a total volume of 25 μl of Taq Plus PCR Master Mix (Qiagen, Valencia, CA, USA) containing 900 nM of the primers and 300 nM of the probes. The cycling conditions were 95°C 10 min, 45× (95°C 15 sec, 60°C 1 min). No-template and non-reverse-transcribed RNAs were used as controls. All reactions were run in triplicate. Signals for αν, β3, NTSR1, PSMA, and PSCA were normalized to RNA input using the signals from TATA binding protein (TBP). The expression differences were calculated by the 2-ΔΔCt method for assessing relative expression.
Table I. Primers and probes used for qRT-PCR.
| Receptor | Primer Sequence (5′ > 3′) | Probe1 (5′ > 3′) |
|---|---|---|
| αν |
F- TTCCCTTCCGGGTAGACG TA R- TGTGCAAAAATAATGCTCTTCGTAT |
TGCTGGATAAACACAAGCAAAGGGAGC |
| β3 |
F- TTTACCACTGATGCCAAGACTCA R- CCGTCATTAGGCTGGACAAT |
CATTGGACGGAAGGCTGGCAG |
| NTSR1 |
F- GGCGCCTCATGTTCTGCTA R- GTGCGTTGGTCACCATGTAGA |
ATGAGCAGTGGACTCCGTTCCTCTATGACTTCT |
| PSCA |
F- CAGGACTACTACGTG GGCAAGA R- CGCTGGCGTTGCACAA |
AACATCACGTGCTGTGACACCGA |
| PSMA |
F- GCTGATAAGCGAGGCATTAGT R- TGCGCGCCCTCCAA |
AGACTTTACCCCGCCGTGGTG |
All probes had a 5′-56-FAM fluorophore and a 36-TAMSp-3′ quencher
Immunohistochemistry
Formalin-fixed tumors were paraffin-embedded and stained by Tricore Reference Laboratory (Albuquerque, NM, USA) using an automated procedure with a Ventana BenchMark XT IHC/ISH Staining Module and polyclonal rabbit anti-human PSCA antibody (Invitrogen, Carlsbad, CA, USA).
Results
Messenger RNA and protein expression levels for integrin ανβ3, NTSR1, PSMA, and PSCA were measured in LNCaP, C4-2, and PC-3 human prostate cancer cell lines using quantitative reverse transcriptase polymerase chain reaction, flow cytometry, and immunohistochemistry. Of particular interest were the differences in the expression levels of these receptors observed between cells cultured in vitro as opposed to cell deposits grown as xenografts in immunocompromised mice in vivo.
Membrane Receptor mRNA Expression in Cells
The baseline mRNA expression of αν, β3, NTSR1, PSMA, and PSCA, was measured using qRT-PCR, in LNCaP, C4-2, and PC-3 human prostate cancer cell lines; these cell types were chosen for their increasing tumorigenic potential (20) in the order LNCaP, C4-2, and PC-3. Figure 1 shows growth curves for these three types of cell deposits grown as xenografts in the flanks of immunocompromised mice. The growth curves suggest that the PC-3 xenografts grow much more rapidly when implanted in the flanks of nude mice, compared to the LNCaP and C4-2 xenografts and, therefore, the PC-3 cell line is much more tumorigenic in this site of implantation. A summary of the mRNA expression is given in Table 2, where the data are reported relative to the least tumorigenic cell line, LNCaP. The LNCaP cell line had the highest amount of PSMA mRNA expression in vitro, but had lower mRNA levels for all of the other receptors, compared to the other two cell lines. The PC-3 cell line, which has the highest tumorigenic potential (20) and was originally collected as a bone metastasis from a patient with CRPC (13) was almost devoid of PSMA mRNA, but showed the highest mRNA expression for αν, NTSR1, and PSCA of all the cell lines. The PC-3 cells also made 5-fold more of the integrin β3 mRNA than the LNCaP cells. These findings imply that an inverse relationship may exist between the tumorigenicity of these cell lines and their PSMA mRNA expression. Conversely, the cellular tumorigenicity appears to positively correlate with the mRNA expression levels of the other three membrane receptors. The C4-2 cell line, a moderately tumorigenic, androgen-independent progeny of LNCaP cells (13), had the highest level of β3 integrin mRNA expression and a 2- to 3-fold greater mRNA expression of αν and NTSR1 than that found for LNCaP cells.
Fig. 1. LNCaP, C4-2, and PC-3 xenograft growth curves.
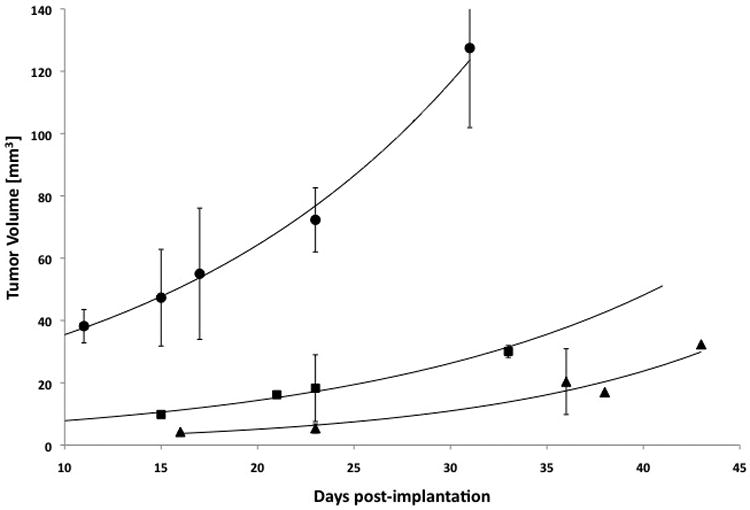
Increase in tumor volume versus days post-implantation of LNCaP, C4-2, or PC-3 cell deposits. Circle = PC-3, Square = C4-2, and Triangle = LNCaP with n = 5, 2, and 3 respectively.
Table II. Receptor mRNA and Protein Expression in Cells and Tumors.
| Cell Line/Xenograft | Cell mRNA Expression | Tissue mRNA Expression | Cell Epitope-Density |
|---|---|---|---|
|
| |||
| PSMA1 | PSMA1 | PSMA2 | |
|
| |||
| LNCaP | 1.00 ± 0.09 | 1.00 ± 0.08 | 105.00 ± 10.0 (104) |
| C4-2 | 0.51 ± 0.04 | 1.37 ± 0.11 | 167.00 ± 28.0 (104) |
| PC-3 | 0.01 ± 0.00 | 0.01 ± 0.00 | 1.42 ± 0.48 (104) |
|
| |||
| PSCA1 | PSCA1 | PSCA2 | |
|
| |||
| LNCaP | 1.00 ± 0.09 | 1.00 ± 0.07 | 0 |
| C4-2 | 0.01 ± 0.00 | 3.81 ± 0.33 | 0 |
| PC-3 | 1.57 ± 0.11 | 1.20 ± 0.10 | 0 |
|
| |||
| αν1 | αν1 | ανβ32 | |
|
| |||
| LNCaP | 1.00 ± 0.02 | 1.00 ± 0.64 | 1.12 ± 0.11 (104) |
| C4-2 | 2.09 ± 0.18 | 3.23 ± 0.05 | 1.17 ± 0.46 (104) |
| PC-3 | 18.61 ± 1.59 | 2.94 ± 0.20 | 0.77 ± 0.02 (104) |
|
| |||
| β3 1 | β3 1 | NA | |
|
|
|||
| LNCaP | 1.00 ± 0.08 | 1.00 ± 0.05 | NA |
| C4-2 | 31.45 ± 2.25 | 1.21 ± 0.01 | NA |
| PC-3 | 5.39 ± 0.17 | 316.60 ± 26.64 | NA |
|
| |||
| NTSR11 | NTSR11 | NTSR13 | |
|
| |||
| LNCaP | 1.00 ± 0.09 | 1.00 ± 0.03 | 67.9 ± 27.1 |
| C4-2 | 2.92 ± 0.23 | 0.05 ± 0.01 | 79.3 ± 15.1 |
| PC-3 | 9.77 ± 0.73 | 88.82 ± 7.61 | 15.9 ± 22.4 |
Expression relative to LNCaP
Receptors per cell
Mean channel fluorescence
NA: not applicable
Membrane Receptor mRNA Expression in Xenografts
In order to determine if the murine microenvironment altered mRNA expression in xenografts as compared to cells, we next measured receptor mRNA levels in LNCaP, C4-2, and PC-3 human prostate cancer cell xenografts grown as subcutaneous tumors in immunocompromised mice. C4-2 and LNCaP xenografts displayed significant PSMA mRNA expression, while the PC-3 xenograft was essentially devoid of PSMA mRNA. The PC-3 xenograft, similar to the cultured PC-3 cells, showed the highest NTSR1 mRNA expression, an ∼88-fold increase with respect to the LNCaP xenograft. The PC-3 xenograft also had ∼316-fold higher β3 mRNA expression and elevated mRNA expression of both αν and PSCA, compared to the LNCaP xenograft. Unexpectedly, the C4-2 xenograft had almost 4-fold higher PSCA mRNA expression, compared to the LNCaP xenograft even though the C4-2 cells lacked PSCA mRNA expression in vitro. Additionally, the C4-2 cell line, which expressed the highest amount of β3 expression, at a ratio of ∼ 30:1 over LNCaP cells, and the second highest amount of NTSR1 mRNA expression, in vitro, lost virtually all of its NTSR1 mRNA expression in the xenograft and also had an ∼ 50-fold reduction in β3 mRNA in vivo.
Comparison of Membrane Receptor mRNA Expression in Cells and Xenografts
To determine the changes in αν, β3, NTSR1, PSMA, and PSCA mRNA expression that may occur when cells grown in a semi-defined medium in vitro are transferred to a more physiological and complex environment in vivo, the qRT-PCR data for the membrane receptors' mRNA expression in xenografts was compared to each corresponding cell line. The results (Table 3) shows that PSMA mRNA decreased by about 50 % in all of the xenografts compared to the same cells grown in culture. A similar decrease in mRNA expression in vivo was evident for PSCA in LNCaP and PC-3 xenografts. However, the C4-2 xenograft displayed PSCA mRNA expression that markedly increased by ∼1450-fold in the xenograft relative to C4-2 cells. The C4-2 xenograft also exhibited modest 1.2 to 2.6-fold increases in αν and NTSR1 mRNA expression relative to C4-2 cells. Large increases in the NTSR1 (615 ×) and β3 (25 ×) mRNA expression were also found in PC-3 xenografts compared to the expression in cells. These data suggested that mRNA expression changed in different environments (in vitro versus in vivo), and added support to the correlation mentioned above between tumorigenicity and expression of cellular mRNA for ανβ3, NTSR1, and PSCA.
Table III. Comparison of mRNA Expression Between Cells and Xenografts.
| Cell Line/Xenograft | Xenograft mRNA Expression Relative to Cells |
|---|---|
|
| |
| PSMA1 | |
|
| |
| LNCaP | 0.30 ± 0.02 |
| C4-2 | 0.63 ± 0.04 |
| PC-3 | 0.55 ± 0.05 |
|
| |
| PSCA1 | |
|
| |
| LNCaP | 0.48 ± 0.03 |
| C4-2 | 1451.15 ± 96.37 |
| PC-3 | 0.37 ± 0.03 |
|
| |
| αν1 | |
|
| |
| LNCaP | 1.69 ± 1.08 |
| C4-2 | 2.61 ± 0.04 |
| PC-3 | 0.27 ± 0.02 |
|
| |
| β31 | |
|
| |
| LNCaP | 0.42 ± 0.02 |
| C4-2 | 0.02 ± 0.00 |
| PC-3 | 24.79 ± 2.09 |
|
| |
| NTSR11 | |
|
| |
| LNCaP | 67.66 ± 2.30 |
| C4-2 | 1.16 ± 0.11 |
| PC-3 | 615.35 ± 20.25 |
mRNA expression relative to the corresponding cell line grown in culture
Membrane Receptor Protein Expression in Cells
It is well known that mRNA expression, as measured both in vitro and in vivo, may not always be representative of actual protein expression levels (21). Therefore, we measured the number of ανβ3, NTSR1, PSMA, and PSCA membrane receptors on the surface of each of the cell lines using flow cytometry and epitope-density calibration particles (See methods). Although the LNCaP cells were found (Table 2) to possess a great number (∼ 1 million) of PSMA proteins per cell, the C4-2 cells displayed the highest PSMA protein expression with ∼ 1.7 million PSMA receptors per cell. Interestingly, although the PC-3 cells were found to contain only low, essentially background, levels of PSMA mRNA, both in vitro and in vivo, a modest number (∼14,000 per PC-3 cell) of PSMA proteins were found on the cell surface. Both C4-2 and LNCaP cell lines expressed ∼11,500 ανβ3 integrins per cell, while the PC-3 cells showed less at ∼7,700 ανβ3 integrins per cell. By measuring the Mean Channel Fluorescence for the NTSR1 protein we found that both LNCaP and C4-2 cells had high expression of NTSR1 on the surface of the cells. The PC-3 cells had ∼ 4.5-fold less NTSR1 protein expression in vitro, compared to LNCaP and C4-2 cells, even though the PC-3 cells had the highest NTSR1 mRNA expression in vitro. The PSCA protein was not expressed on the surface of any of the three cell lines in vitro.
Immunohistochemistry for PSCA in Xenografts
Even though the PSCA protein could not be detected (Table 2) on the surfaces of our cell lines, the mRNA expression data from the xenografts (Table 3) indicated that PSCA might be actually expressed in vivo, particularly for the C4-2 cell line where these xenografts made more than 1400 times as much mRNA as the cells. For this reason, PSCA protein expression was determined by immunohistochemistry (IHC) in LNCaP, C4-2, and PC-3 xenografts generated in immunocompromised mice. Although PSCA protein was undetectable on the surfaces of cells cultured in vitro by the flow cytometric technique (Table 2), punctate staining for PSCA was observed within the tissue cells by IHC in all xenografts (Figure 2A-C). The staining intensity for PSCA in these xenografts was comparable to the intensity found in human prostate cancer tissue used as a positive control (Figure 2F), while there was a complete absence of PSCA staining, either in the mouse splenic tissue (Figure 2E), or human prostate cancer tissue incubated in the absence of the primary antibody (Figure 2D), which were used as negative controls. The staining for PSCA observed in the IHC of the xenografts was the same type of punctate staining as reported by others in human tumors, and in LAPC-4 cells that are known to highly-express surface PSCA (22).
Fig. 2. Immunohistochemical staining for PSCA in human prostate cancer xenografts in immunocompromised mice and in human prostate cancer tissue.
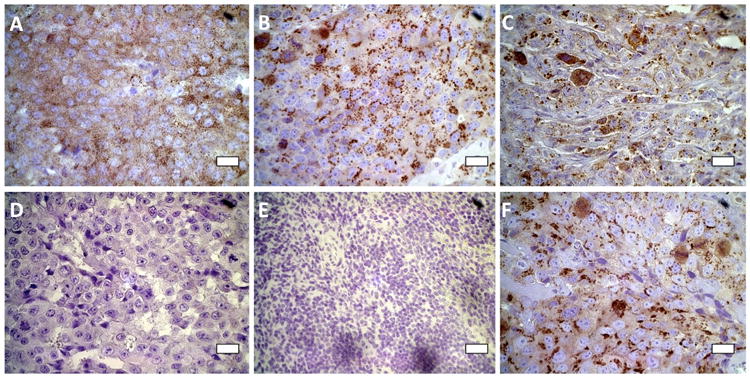
(A) LNCaP xenograft; (B) C4-2 xenograft; (C) PC-3 xenograft; (D) human malignant prostate tissue in the absence of primary antibody [negative control]; (E) mouse spleen [negative control]; (F) human malignant prostate tissue in the presence of primary antibody [positive control]. Scale bars are 20 μm.
Taken together, the mRNA and protein data emphasize that regulation of mRNA expression of the membrane bound and potential target proteins ανβ3, NTSR1, PSMA, and PSCA in prostate cancer cells with different tumorigenic potential can be influenced by factors of the microenvironment, such as in murine xenotransplants. In addition, correlations appear to exist between membrane receptor expression signatures and tumorigenicity, but mRNA expression levels for these proteins do not translate directly to equivalent expression levels of membrane bound protein. These trends are graphically summarized in Figure 3.
Fig. 3. Trends of membrane receptor mRNA and protein expression in human prostate cancer cells and in murine xenografts.
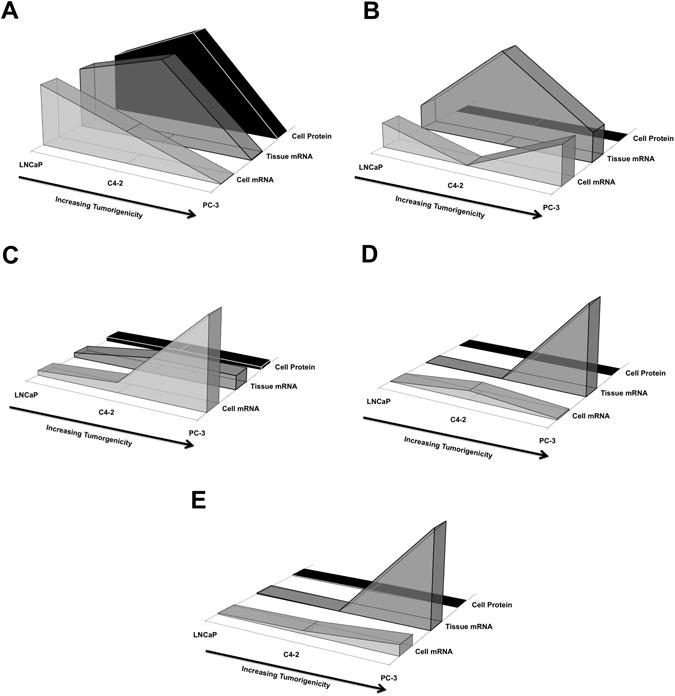
Expression of membrane receptors PSMA (A), PSCA (B), integrin αν (C), integrin β3 (D), and NTSR1 (E) for human prostate cancer cells of increasing tumorigenic potential, in the order: LNCaP, C4-2, and PC-3. Messenger RNA expression levels for cells cultured in vitro (front shapes) and as xenografts in vivo (middle shapes), and protein levels (back shapes) are shown normalized to LNCaP cells or xenografts.
Discussion
Three main findings are reported in this study that are of importance for research on improved prostate cancer imaging and therapeutic targeting agents, as exemplified by our previously reported functionalized iron oxide nanoparticles (23). First, our findings indicate the necessity of verifying the presence of target proteins at the cell surface, as the level of mRNA expression does not necessarily translate into protein expression levels. This discrepancy was evident for PSMA expression in PC-3 cells. We have confirmed that PC-3 cells express little or no PSMA (24), even though these cells represent the most tumorigenic and advanced prostate cancer cell line examined here. On the other hand, in advanced CRPC in humans, PSMA expression increased markedly with tumor grade, stage, and after androgen-deprivation therapy (25). Our data show a 100-fold decrease in the expression of PSMA mRNA in PC-3 cells and xenografts compared with the LNCaP and C4-2 lines. This decrease in mRNA corresponded to a similar ∼100-fold decrease (to ∼14,000 from more than 1 million) in the number of PSMA receptors per cell (Figure 3A and Table 2). Further discrepancies were observed for PSCA, for which even though mRNA was expressed in all of the cells analyzed, and no protein could be detected on their cell surfaces (Figure 3B and Table 2), was yet expressed in xenografts (Figure 2A-C). We found that ανβ3 protein expression was comparable in all three of the cell lines, although both αν and β3 mRNA levels appeared to increase with tumorigenicity in the three different cell lines and xenografts. Similar trends were observed for NTSR1 in C4-2 and PC-3 cells (Figure 3E), indicating that the comparative rate of protein translation for these surface markers can greatly differ in prostate cancer cells of various origins.
A second important finding is the discordant expression of some of the surface markers under investigation between cells grown in culture and as xenografts in immunocompromised mice. Marked differences were found for integrin β3, NTSR1, and PSCA (Figure 3 and Table 3). Most prominently, PSCA mRNA expression was dramatically enhanced in C4-2 xenografts (Figure 3B and Table 3) but the protein was not detectable on the surface of the parent cell line. The presence of the PSCA protein in C4-2 and other xenografts, verified by IHC, resembled PSCA expression in human tissues (Figure 2). PSCA is a glycosylphosphatidylinositol (GPI)-anchored membrane antigen that has been reported to be over-expressed in both primary and metastatic prostate cancer lesions (18,26,27). Since we measured PSCA mRNA expression both in vitro and in vivo in all cell lines, one possible explanation for the lack of PSCA protein in vitro could be that PSCA protein is not translated in the absence of the extracellular matrix (ECM). GPI-anchors are known to be added to the C-terminus of peptides as a post-translational modification (28), and thus, PSCA may not have the proper GPI-anchor attached in vitro. Although a detailed understanding of PSCA regulation is still elusive, our data suggest that PSCA expression is affected by cell type and ECM dependent contact.
Integrin ανβ3 has been proposed as a neovasculature-targeting motif for diagnostics and therapeutics due to its over-expression on newly formed vasculature within tumors (16,29-32). In this study, both integrin subunits experienced an induction of expression in vivo in the more tumorigenic cell types, i.e. C4-2 and PC-3, potentially as a consequence of cell-cell and cell-ECM interactions. Similarly, relative strong inductions in the in vivo setting were observed for NTSR1 in LNCaP and PC-3 cells (Figure 3E and Table 3). The NTSR1 receptor is over-expressed in numerous types of solid tumors and NTSR1 receptor binding to neurotensin (NT) has been reported to increase proliferation of several types of cancer cells, including prostate cancer cells (17,33). Further, NT functions via autocrine, paracrine, and endocrine actions in prostate cancer tissues (34,35). Consequently, a plausible explanation for the observed induction of NTSR1 in xenografts could be due to autocrine NT stimulation.
A final important finding is that a relationship appears to exist between membrane receptor signatures and tumorigenicity. We found that PSMA mRNA and protein expression levels tended to be inversely related to tumorigenic potential, both in vitro and in vivo. Additionally, ανβ3 tended to increase with tumorigenic potential both in vitro and in vivo (Figure 3C-D). It is possible that the degree of ανβ3 expression is not only dependent on the extent of vascularization within tumors, but also on the tumorigenic potential of the cells. We also found that NTSR1 mRNA expression increased with increasing tumorigenic potential both in vitro and in vivo yet, NTSR1 protein expression was inversely related to tumorigenic potential in vitro, reflecting our first main finding above. In the future, it may also be worthwhile to compare membrane receptor expression changes that may or may not occur when the cell deposits are instead implanted in the prostate or bone of immunocompromised mice.
Taken together, our data demonstrate that the membrane receptor expression profiles are altered with analogous changes in tumorigenic potential and that these alterations may comprise signatures of the tumorigenic state. Moreover, these membrane receptor signatures were altered for in vitro and in vivo models. We conclude that targeting nanoparticles, diagnostics, and therapeutics with multiple antibodies or peptides against PSMA as well as ανβ3, NTSR1, and/or PSCA may be more beneficial in diagnosing and treating early stage prostate cancer and CRPC than PSMA targeting alone. In addition, a major finding is that cell lines that do not express certain receptors, such as PSCA, in vitro, may very well express these receptors in vivo and prove to be useful receptors for targeting novel agents in humans.
Conclusions
PSMA is the membrane receptor most frequently used for targeting prostate cancer cells (23,30,36-38), and we conclude that LNCaP and the castration resistant and more tumorigenic C4-2 cells are ideal cell models for PSMA directed targeting, as these cell lines display relatively high and persistent PSMA protein expression in vitro and in vivo. However, we provide evidence that additional targeting motifs exist that could increase the specificity and efficacy of imaging and treatment schemes, as shown by the expression of ανβ3, NTSR1, and PSCA in relevant cell and xenograft models of prostate cancer. In fact, co-targeting strategies may be necessary in light of the fact that membrane receptor signatures may change over time as the tumor progresses and that intra-tumoral heterogeneity may lead to variability in expression of any single membrane receptor, thereby hampering efficacy. Furthermore, we found that membrane receptor signatures change not only with alterations in tumorigenicity but are also modified in in vitro and in vivo models. We suggest that designing targeted diagnostics and/or therapeutics using cell models such as LNCaP, C4-2, and PC-3 should ideally include up-front in vivo measurements of the target membrane receptors, which may reveal optimal models under physiologically relevant conditions.
This study provides a novel comparison of expression signatures of prominent membrane receptors for prostate cancer targeting using widely used prostate cancer cells grown in vitro and in vivo. Knowledge reported herein should be helpful in guiding the development of targeting strategies for imaging and therapeutic agents using membrane receptor signatures rather than single membrane-bound targets. This approach should in turn overcome the difficulties often encountered when translating in vitro applications to pre-clinical models and when transitioning such applications towards clinical use.
Acknowledgments
This research was supported by the National Institutes of Health grant 5RO1CA123194 to L.O.S. We acknowledge the use of the Keck-UNM Genomics Resource, a facility supported by the WM Keck Foundation, the State of New Mexico and the UNM Cancer Research and Treatment Center. We thank the UNM Flow Cytometry Shared Resource Center, which is supported by the University of New Mexico Health Sciences Center and the University of New Mexico Cancer Center. We also thank Tricore Reference Laboratory, Albuquerque, NM for their services in immunohistochemistry, and the UNM Department of Biochemistry and Molecular Biology for their vital administrative support.
Support: National Institutes of Health grant 5RO1CA123194
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011 doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Sardana G, Dowell B, Diamandis EP. Emerging biomarkers for the diagnosis and prognosis of prostate cancer. Clin Chem. 2008;54(12):1951–1960. doi: 10.1373/clinchem.2008.110668. [DOI] [PubMed] [Google Scholar]
- 3.Gupta A, Roobol MJ, Savage CJ, Peltola M, Pettersson K, Scardino PT, Vickers AJ, Schroder FH, Lilja H. A four-kallikrein panel for the prediction of repeat prostate biopsy: data from the European Randomized Study of Prostate Cancer Screening in Rotterdam, Netherlands. Br J Cancer. 2010;103(5):708–714. doi: 10.1038/sj.bjc.6605815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Studer UE, Collette L. What can be concluded from the ERSPC and PLCO trial data? Urol Oncol. 2010;28(6):668–669. doi: 10.1016/j.urolonc.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Rabbani F, Stroumbakis N, Kava BR, Cookson MS, Fair WR. Incidence and clinical significance of false-negative sextant prostate biopsies. J Urol. 1998;159(4):1247–1250. [PubMed] [Google Scholar]
- 6.Zarour L, Alumkal J. Emerging therapies in castrate-resistant prostate cancer. Curr Urol Rep. 2010;11(3):152–158. doi: 10.1007/s11934-010-0104-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Lorenzo G, Buonerba C, Autorino R, De Placido S, Sternberg CN. Castration-resistant prostate cancer: current and emerging treatment strategies. Drugs. 2010;70(8):983–1000. doi: 10.2165/10898600-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 8.Peng XH, Qian X, Mao H, Wang AY, Chen ZG, Nie S, Shin DM. Targeted magnetic iron oxide nanoparticles for tumor imaging and therapy. Int J Nanomedicine. 2008;3(3):311–321. doi: 10.2147/ijn.s2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afnan J, Tempany CM. Update on prostate imaging. Urol Clin North Am. 2010;37(1):23–25. doi: 10.1016/j.ucl.2009.11.009. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jadvar H. Molecular imaging of prostate cancer: a concise synopsis. Mol Imaging. 2009;8(2):56–64. [PMC free article] [PubMed] [Google Scholar]
- 11.Chellat F, Merhi Y, Moreau A, Yahia L. Therapeutic potential of nanoparticulate systems for macrophage targeting. Biomaterials. 2005;26(35):7260–7275. doi: 10.1016/j.biomaterials.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 12.Adair JH, Parette MP, Altinoglu EI, Kester M. Nanoparticulate alternatives for drug delivery. ACS Nano. 2010;4(9):4967–4970. doi: 10.1021/nn102324e. [DOI] [PubMed] [Google Scholar]
- 13.Sobel RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines--part 1. J Urol. 2005;173(2):342–359. doi: 10.1097/01.ju.0000141580.30910.57. [DOI] [PubMed] [Google Scholar]
- 14.Sobel RE, Sadar MD. Cell lines used in prostate cancer research: a compendium of old and new lines--part 2. J Urol. 2005;173(2):360–372. doi: 10.1097/01.ju.0000149989.01263.dc. [DOI] [PubMed] [Google Scholar]
- 15.Emonds KM, Swinnen JV, Mortelmans L, Mottaghy FM. Molecular imaging of prostate cancer. Methods. 2009;48(2):193–199. doi: 10.1016/j.ymeth.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 16.McCabe NP, De S, Vasanji A, Brainard J, Byzova TV. Prostate cancer specific integrin alphavbeta3 modulates bone metastatic growth and tissue remodeling. Oncogene. 2007;26(42):6238–6243. doi: 10.1038/sj.onc.1210429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swift SL, Burns JE, Maitland NJ. Altered expression of neurotensin receptors is associated with the differentiation state of prostate cancer. Cancer Res. 2010;70(1):347–356. doi: 10.1158/0008-5472.CAN-09-1252. [DOI] [PubMed] [Google Scholar]
- 18.Saeki N, Gu J, Yoshida T, Wu X. Prostate stem cell antigen: a Jekyll and Hyde molecule? Clin Cancer Res. 2010;16(14):3533–3538. doi: 10.1158/1078-0432.CCR-09-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL, Pathak S, von Eschenbach AC, Chung LW. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54(10):2577–2581. [PubMed] [Google Scholar]
- 20.Aalinkeel R, Nair MP, Sufrin G, Mahajan SD, Chadha KC, Chawda RP, Schwartz SA. Gene expression of angiogenic factors correlates with metastatic potential of prostate cancer cells. Cancer Res. 2004;64(15):5311–5321. doi: 10.1158/0008-5472.CAN-2506-2. [DOI] [PubMed] [Google Scholar]
- 21.Gry M, Rimini R, Stromberg S, Asplund A, Ponten F, Uhlen M, Nilsson P. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu Z, Thomas G, Yamashiro J, Shintaku IP, Dorey F, Raitano A, Witte ON, Said JW, Loda M, Reiter RE. Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19(10):1288–1296. doi: 10.1038/sj.onc.1203426. [DOI] [PubMed] [Google Scholar]
- 23.Serda RE, Adolphi NL, Bisoffi M, Sillerud LO. Targeting and cellular trafficking of magnetic nanoparticles for prostate cancer imaging. Mol Imaging. 2007;6(4):277–288. [PubMed] [Google Scholar]
- 24.Laidler P, Dulinska J, Lekka M, Lekki J. Expression of prostate specific membrane antigen in androgen-independent prostate cancer cell line PC-3. Arch Biochem Biophys. 2005;435(1):1–14. doi: 10.1016/j.abb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Chang SS, Reuter VE, Heston WD, Hutchinson B, Grauer LS, Gaudin PB. Short term neoadjuvant androgen deprivation therapy does not affect prostate specific membrane antigen expression in prostate tissues. Cancer. 2000;88(2):407–415. [PubMed] [Google Scholar]
- 26.Raff AB, Gray A, Kast WM. Prostate stem cell antigen: a prospective therapeutic and diagnostic target. Cancer Lett. 2009;277(2):126–132. doi: 10.1016/j.canlet.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhigang Z, Wenlu S. Complete androgen ablation suppresses prostate stem cell antigen (PSCA) mRNA expression in human prostate carcinoma. Prostate. 2005;65(4):299–305. doi: 10.1002/pros.20290. [DOI] [PubMed] [Google Scholar]
- 28.Lakhan SE, Sabharanjak S, De A. Endocytosis of glycosylphosphatidylinositol-anchored proteins. J Biomed Sci. 2009;16:93. doi: 10.1186/1423-0127-16-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong H, Zhang Y, Sun J, Cai W. Positron emission tomography imaging of prostate cancer. Amino Acids. 2010;39(1):11–27. doi: 10.1007/s00726-009-0394-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stollman TH, Ruers TJ, Oyen WJ, Boerman OC. New targeted probes for radioimaging of angiogenesis. Methods. 2009;48(2):188–192. doi: 10.1016/j.ymeth.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Schmieder AH, Winter PM, Caruthers SD, Harris TD, Williams TA, Allen JS, Lacy EK, Zhang H, Scott MJ, Hu G, Robertson JD, Wickline SA, Lanza GM. Molecular MR imaging of melanoma angiogenesis with alphanubeta3-targeted paramagnetic nanoparticles. Magn Reson Med. 2005;53(3):621–627. doi: 10.1002/mrm.20391. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Wang F, Chen X. Integrin alpha(v)beta(3)-Targeted Cancer Therapy. Drug Dev Res. 2008;69(6):329–339. doi: 10.1002/ddr.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almeida TA, Rodriguez Y, Hernandez M, Reyes R, Bello AR. Differential expression of new splice variants of the neurotensin receptor 1 gene in human prostate cancer cell lines. Peptides. 2010;31(2):242–247. doi: 10.1016/j.peptides.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Carraway RE, Plona AM. Involvement of neurotensin in cancer growth: evidence, mechanisms and development of diagnostic tools. Peptides. 2006;27(10):2445–2460. doi: 10.1016/j.peptides.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 35.Carraway RE, Hassan S. Neurotensin receptor binding and neurotensin-induced growth signaling in prostate cancer PC3 cells are sensitive to metabolic stress. Regul Pept. 2007;141(1-3):140–153. doi: 10.1016/j.regpep.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 36.Wang W, Mo ZN. Advances in prostate-specific membrane antigen targeted therapies for prostate cancer. Zhonghua Nan Ke Xue. 2010;16(6):547–551. [PubMed] [Google Scholar]
- 37.Elsasser-Beile U, Buhler P, Wolf P. Targeted therapies for prostate cancer against the prostate specific membrane antigen. Curr Drug Targets. 2009;10(2):118–125. doi: 10.2174/138945009787354601. [DOI] [PubMed] [Google Scholar]
- 38.Bander NH. Technology insight: monoclonal antibody imaging of prostate cancer. Nat Clin Pract Urol. 2006;3(4):216–225. doi: 10.1038/ncpuro0452. [DOI] [PubMed] [Google Scholar]


