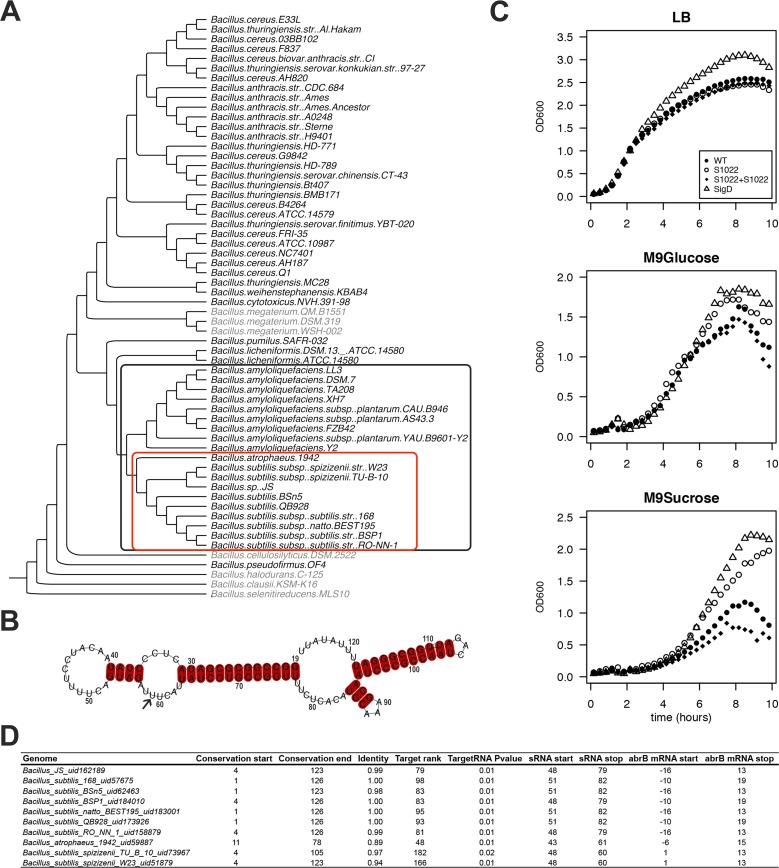
Fig 1. The RnaC/S1022 growth phenotype is linked to the evolutionary target prediction of abrB.
A) Phylogenetic tree of Bacillus genomes. The tree was constructed based on an alignment of rpoB (present in 60 of the 62 genomes; except for two Bacillus coagulans genomes for which no significant rpoB nBLAST hits were found). The outer (black) box indicates 19 genomes in which RnaC/S1022 is present. The inner (red) box indicates 10 genomes in which the predicted RnaC/S1022-abrB interaction is consistently observed. A significant nBLAST hit for AbrB was not obtained for the species shaded in grey. B) LocARNA structural conservation alignment of RnaC/S1022 based on the sequence published by Schmalisch et al. [28]. The alignment includes RnaC/S1022 sequences from genomes in which the RnaC/S1022-abrB interaction is consistently observed (marked in the red box in panel A), except the RnaC/S1022 from B. atrophaeus (see also S1 Fig.). Numbers indicate the coordinates of S1022 defined by Nicolas et al. [9]. The arrow highlights the uracil base that is required for the interaction with abrB. C) Growth curves of parental strain 168trp+, ΔRnaC/S1022, ΔsigD, and the ΔRnaC/S1022 amyE::RnaC/S1022 complemented strain grown on LB, M9G or M9S. Each experiment was repeated at least three times in 96-well plates and shake flasks. Averages from triplicates from a representative 96-well plate experiment are shown. The OD600 was monitored every 10 min. One in two time-points were plotted. D) Overview of the predicted RnaC/S1022-abrB interaction. “Genome”, name of the bacterium in which the interaction was predicted. “Conservation start”, the base of the RnaC/S1022 sequence as defined by Nicolas et al. [9] where the significant nBLAST hit starts. “Conservation end”, end coordinate of the significant nBLAST hit. “Identity”, fraction of identity of the conserved RnaC/S1022 sequence compared to RnaC/S1022 of B. subtilis 168. “Target Rank” the ranking of the abrB target in the predicted RnaC/S1022 targets for the respective genome. “TargetRNA P value” TargetRNA_v1 prediction P-value. “sRNA_start”, start coordinate of RnaC/S1022 homologue in the predicted target interaction. “sRNA_stop”, end coordinate of RnaC/S1022 homologue in the predicted target interaction. “abrB mRNA_start”, start coordinate of abrB in the predicted target interaction relative to its start codon. “abrB mRNA_stop”, end coordinate of abrB in the predicted target interaction relative to its start codon.