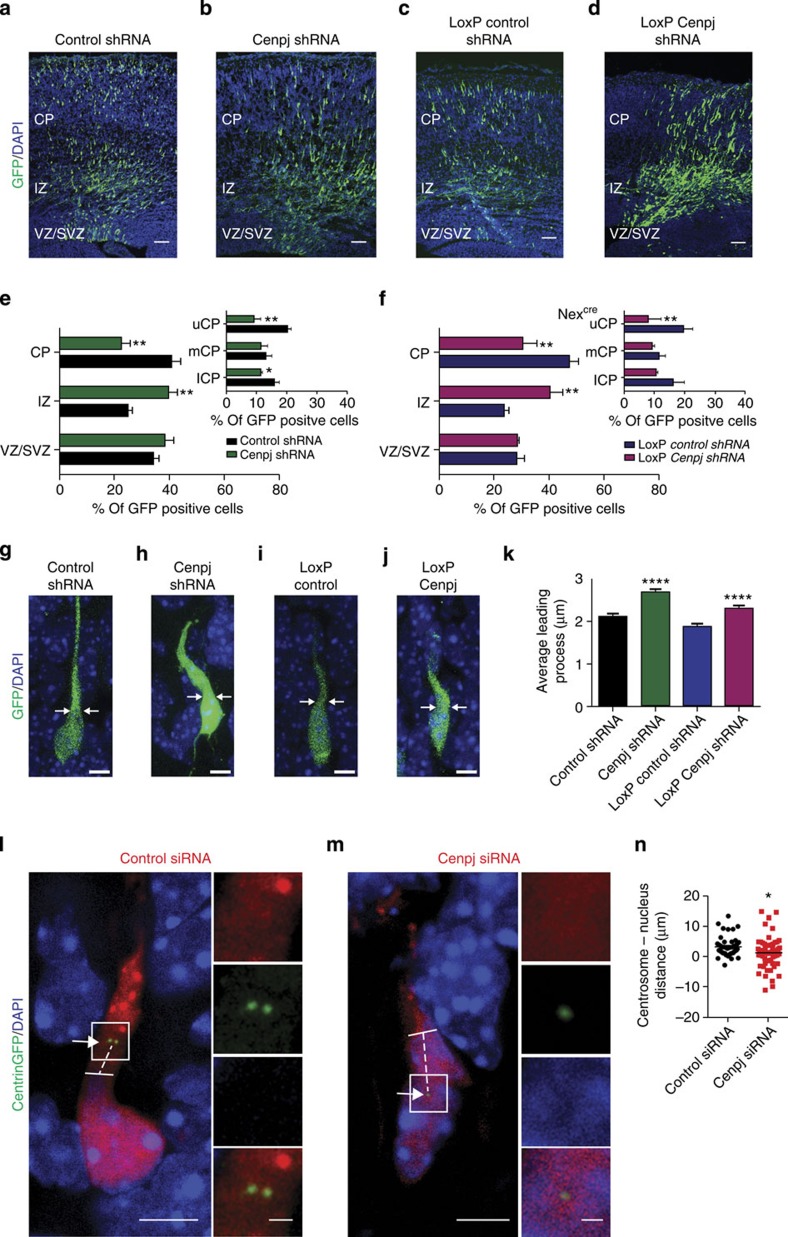
Figure 3. Silencing Cenpj alters neuronal migration and morphology.
(a–d) Analysis of radial migration by GFP labelling of cortices 3 days after in utero co-electroporation at E14.5 with GFP and control shRNA (a), Cenpj shRNA (b). pCALSL control shRNA (c) and conditionally activable pCALSL_Cenpj shRNA (d). pCALSL control and_Cenpj shRNA were electroporated in Nex-cre embryos. Scale bar, 50 μm. (e,f) Quantification of the migration defect of neurons silenced for Cenpj in VZ progenitors (e) or in Nex+ neurons (f) by measuring the percentage of GFP+ cells that have reached the different zones of the cortex 3 days after electroporation. Cells silenced for Cenpj at both progenitor and neuronal stages accumulate more in the IZ and less in the upper CP than control cells. Student’s t-test *P<0.05, **P<0.01. (g–j) Cenpj-depleted neurons silenced at both progenitor (h) and neuronal stages (j) display abnormal enlargement of the leading process 3 days after electroporation compared with control neurons (g,i). Scale bar, 5 μm. (k) Quantification of the thickness of the leading process performed 3 days after electroporation, as described in the Methods section. Cenpj-depleted neurons have a thicker leading process. Three embryos analysed for each condition; control shRNA, n=177 cells; Cenpj shRNA, n=200 cells. Student’s t-test ****P<0.0001. (l,m) Analysis of the distance between the centrosome and the nucleus in migrating cortical neurons 3 days after in utero co-electroporation at E14.5 of control or Cenpj shRNAs together with pClG2-Centrin2-Venus for labelling of the centrioles and pCMV-RFP construct to mark electroporated cells. The nuclei are labelled with 4',6-diamidino-2-phenylindole (DAPI). Insets show higher magnification of the boxed areas with separate colour channels. Arrows point to the centrosome, dashed lines mark the distance between the centrosome and the tip of the nucleus, marked by a full line. Scale bar, 3 μm (main panels) and 1 μm (insets). (n) Quantification of the distance between the centrosome and the nucleus in migrating cortical neurons. Negative values correspond to centrosomes located below the tip of the nucleus. Cenpj-depleted neurons present more negative centrosome–nucleus distances than control neurons. Three embryos analysed for each condition; control shRNA, n=47 cells; Cenpj shRNA, n=60. Student’s t-test *P<0.05.