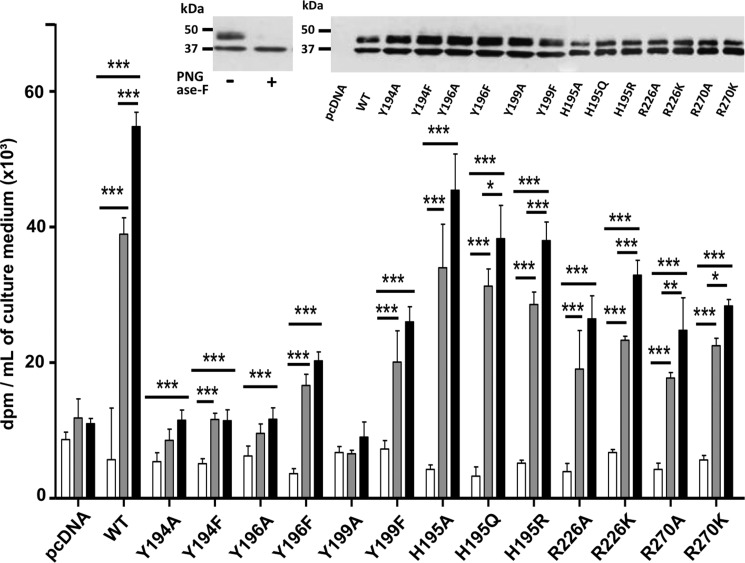
FIGURE 2.
Effect of wild-type and mutated hβ4GalT7 expression on GAG chains primed from 4-MUX in CHOpgsB-618 cells. Cells were transiently transfected with wild-type (WT) or mutated hβ4GalT7 cDNA or with empty vector (pcDNA), and GAG chains synthesis was quantified by scintillation counting following Na2[35SO42−] incorporation, using 0 (white bars), 5 (gray bars), and 10 μm (black bars) 4-MUX. Immunoblot analyses of the protein expression level in CHOpgsB-618 cells transfected with the vector coding for the wild-type or mutated hβ4GalT7 are shown as the inset. The enzyme was identified at the band of ∼35 kDa, whereas the upper band corresponding to ∼39 kDa band could be attributed to the N-glycosylated enzyme as demonstrated by its disappearance upon addition of peptide N-glycosidase F (PNGase F) (left panel). Both bands intensities were used to quantify the total protein expression level (ImageJ software). The immunoblot analysis indicates that the mutated enzymes were all expressed at a comparable level to that of the wild-type hβ4GalT7 (right panel). Data are mean ± S.E. of three independent experiments performed in triplicate. Statistical analysis was carried out by the Student's t test with *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus GAG synthesis in the absence of 4-MUX.