FIGURE 3.
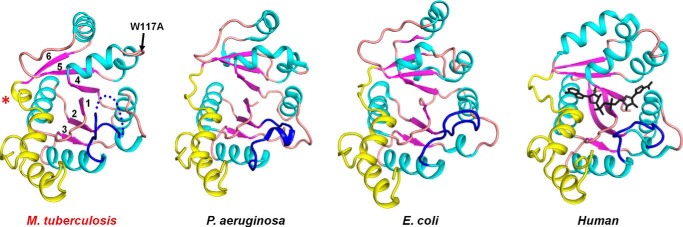
Structural topology of MtNadD, E. coli NadD (PDB code 1K4K), P. aeruginosa NadD (PDB code 1YUL), and human NMNAT (PDB code 1KKU). The overall topology of MtNadD is highly similar to other representatives of the bacterial NadD family illustrated by examples from Gram-negative and Gram-positive species and to a distantly homologous human NMNAT. For comparison, all apo forms of the protein family were used. The position of the active site is indicated by the NaAD product (black) from PDB code 1KQO superimposed on the human enzyme. Secondary elements of the core Rossman fold are depicted in cyan helices, purple strands, and an orange coil. The variable C-terminal domain is in yellow. The flexible loop that has been implicated in NaMN binding is in blue. Disordered segments are depicted with dotted traces. The asterisk indicates the 310 helix η1, a unique feature of MtNadD, replacing an extended coil in all other characterized NaMN/NMN adenylyltransferases. When present, the product molecule is shown in a stick representation.
