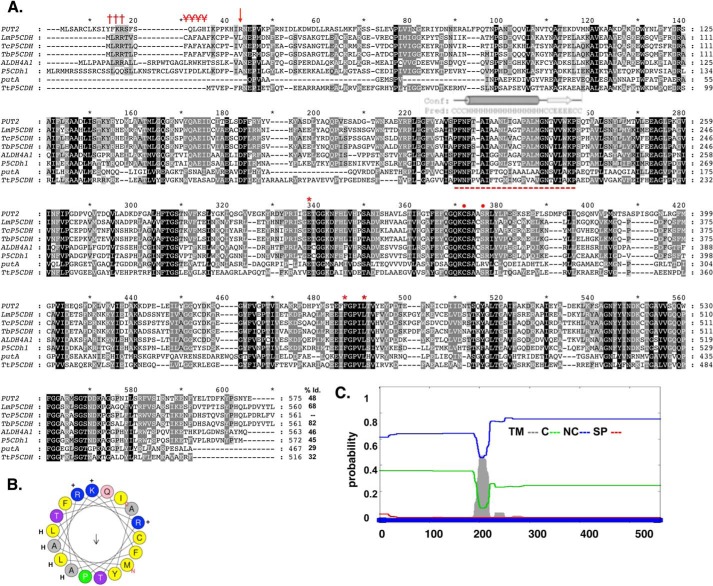
FIGURE 1.
In silico analysis of the deduced amino acid sequence for TcP5CDH. A, alignment of primary sequences for P5CDHs from different species. The aligned sequences correspond to orthologous proteins from H. sapiens (ALDH4A1), D. melanogaster (P5CDh1), T. cruzi (TcP5CDH), T. brucei (TbP5CDH), L. major (LmP5CDH), S. cerevisiae (PUT2), E. coli (putA), and T. thermophilus (TtP5CDH). Accession code numbers are detailed under “Experimental Procedures.” The alignment was performed using the ClustalW method (default parameters) (41). Protein signatures found in the mTP (Met1–Ser18) indicating the MLRR (†††) and alanine-rich FAFAYA (¥¥¥) domains and the putative cleavable site (arrow) of TcP5CDH. The predicted trans-membrane region (Phe200–Trp221) is highly conserved among trypanosomatid sequences (- - -), as are the conserved catalytic residues Glu302, Phe387, Leu441 (stars), and Cys336, Ser340 (circles), as shown by previous structural data. B, α-helical structure presented in mTP region using the heliQuest software. The resulting model depicts the positive (+) (Arg3, Arg4, and Lys16) and hydrophobic (H) (Leu2, Leu6, Ala9, Ala11, and Ala13) residues and the putative Thr17 (arrow) cleavable site. C, bioinformatic analysis for the transmembrane region found for TcP5CDH. The complete amino acid sequence for TcP5CDH (561 residues) was used as the input sequence for the Phobius predictor tool. The graphic layout shows the probability values (0–1) for the transmembrane (TM) domains in gray, cytoplasmic (C) in green, noncytoplasmic (NC) in blue and signal peptide (SP) in red.