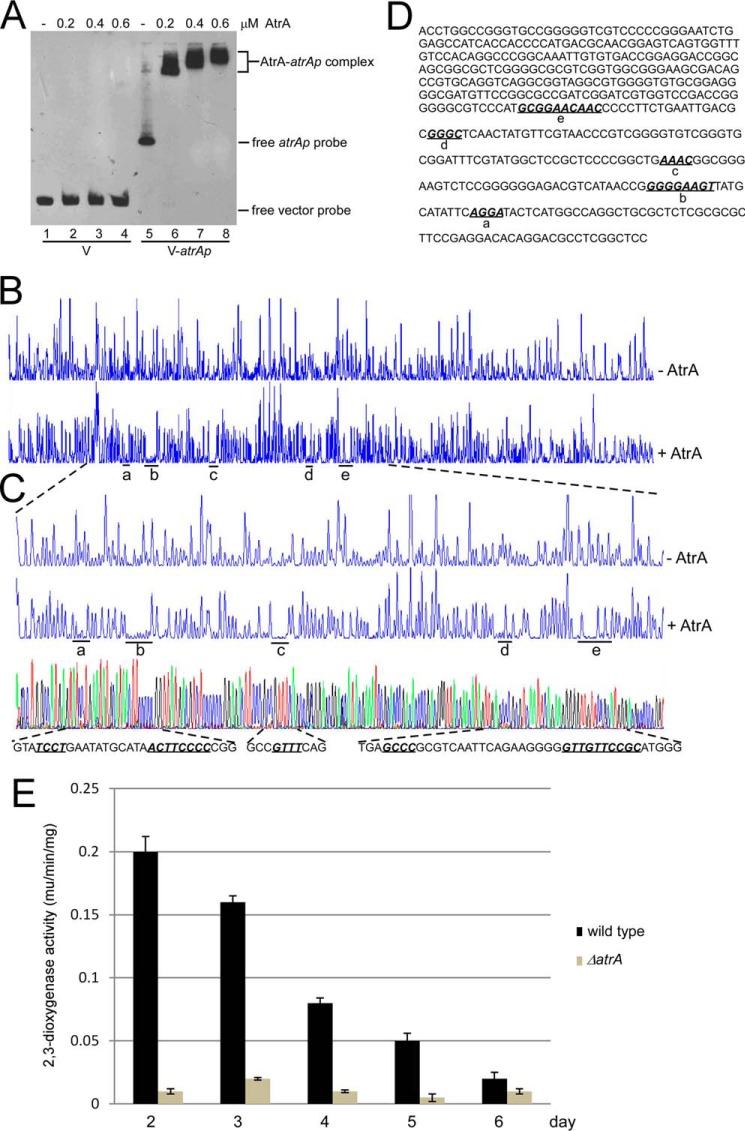
FIGURE 7.
Binding of AtrA to its own promoter (atrAp). A, AtrA binds to atrAp. Biotin-labeled atrAp (V-atrAp) was used as a probe, and biotin-labeled void vector (V) was the negative control. Gradient concentrations of AtrA were added for EMSA. B, DNase I footprinting assay of AtrA binding sites on atrAp. FAM-labeled atrAp was used as the probe with 1 μm AtrA. The protected regions were labeled as a–e, respectively. C, binding sequence determination of AtrA on atrAp by DNase I footprinting assays. D, the binding sequences of AtrA on atrAp as determined by DNase I footprinting assays. The five binding sites are shown in bold italics and are underlined. E, AtrA positively regulates atrAp activity. The wild type and the ΔatrA mutant were transformed with pIPP1-atrAp (atrAp-xylE) and cultured in YEME medium. Cells were collected and disrupted by sonication every day, and 2,3-dioxygenase activity was measured. The data were from three independent experiments. S.D. bars are shown.