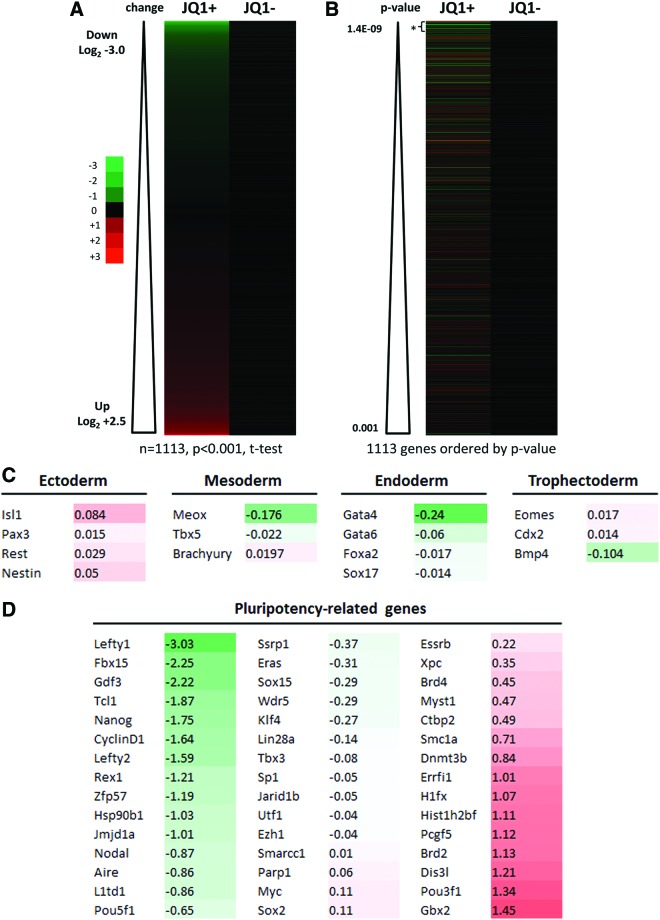
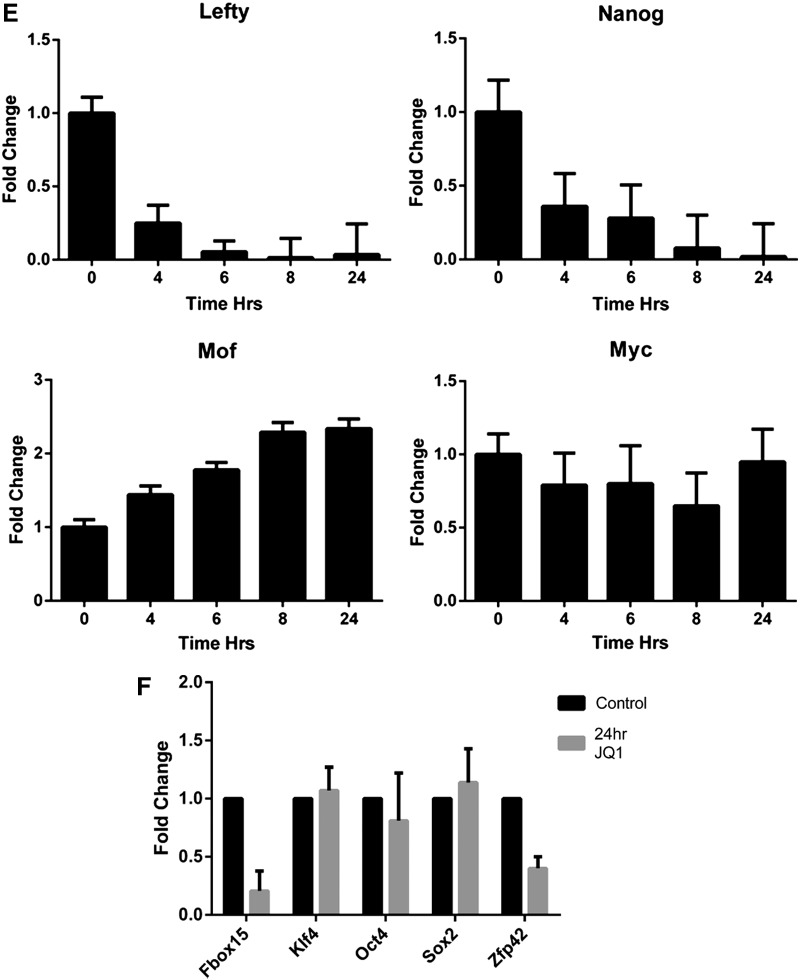
FIG. 2.
JQ1 induces transcriptional deregulation of genes. Microarray profiling was performed using the Illumina platform to assess the transcriptional effects of 1 μM JQ1 and (−)-JQ1 at 16 h on ESCs. (A) Heatmaps (derived from the analysis of 4 replicates) showing expression level changes for the most deregulated genes (n=1,113 genes at t-test significance level of P<0.001) ordered by degree of deregulation (green=up; red=down); (B) heatmaps of the most significantly deregulated genes ordered by P values (JQ1 versus untreated control, two-tailed t-test) with analysis of the 50 most deregulated genes (*) revealing a predominant downregulatory effect compared with the rest (mean log2−0.6084 versus −0.0026, respectively, P=9.6×10−11, two-tailed t-test); (C) effect of JQ1 on transcription of germ layer-specific genes; (D) effect of JQ1 on selected pluripotency-related genes; (E) confirmatory qRT-PCR analysis of cDNA samples prepared from ESCs treated with 0.5 μM for up to 24 h was performed for Nanog, Lefty1, Mof, and Myc; (F) quantification of mRNA transcript levels for Fbox15, klf4, Oct4, Sox2, and Zfp42 was performed by qRT-PCR (data are representative of at least three independent experiments performed in triplicate±SD). Color images available online at www.liebertpub.com/scd