Figure 4.
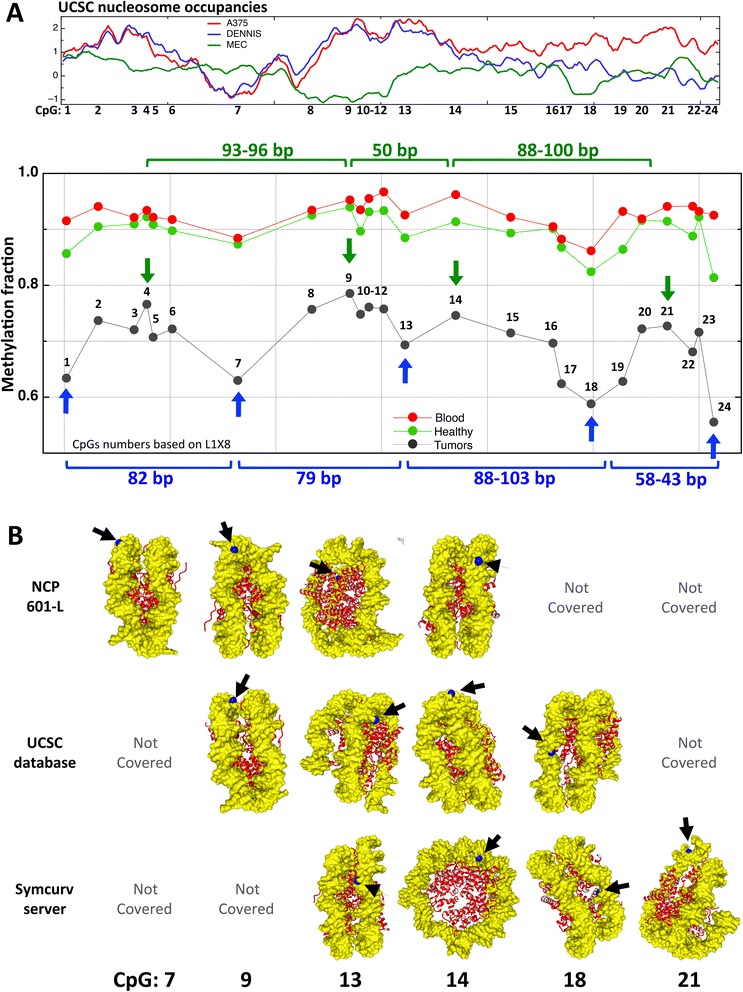
Summary presentations of the relative location of hypomethylated CpG sites at five X-linked LINE-1 loci. (A) The upper panel represents the predicted averaged nucleosomal occupancy of all five X-linked LINE-1s, based on the UCSC track: regulation, nucleosome occupancy [42]. The high peaks of red and blue lines (experimental results designated as A375 and DENNIS at the UCSC, respectively) correspond to a high probability of nucleosome occupancy, while the green peaks (designated as MEC) correspond to a low probability of nucleosome occupancy. In the lower panel, graphs represents the average methylation of all five X-linked LINE-1 loci for the six sequenced tumor samples (black), for their normal tissue controls (green), and for four blood controls (red). Vertical blue and green arrows represent the relatively hypo- and hypermethylated peaks (CpG numbering is according to L1X8). (B) The predicted localization of a particular cytosine carbon based on the three models of LINE-1 DNA around the histone octamers (as described in methods). The cytosine carbons in question are labeled in blue, histones are in red, and DNA is in yellow. The number of the CpGs whose C is labeled is shown at the lower part of the graph. NCP: nucleosome core particle; UCSC: University of California, Santa Cruz.
