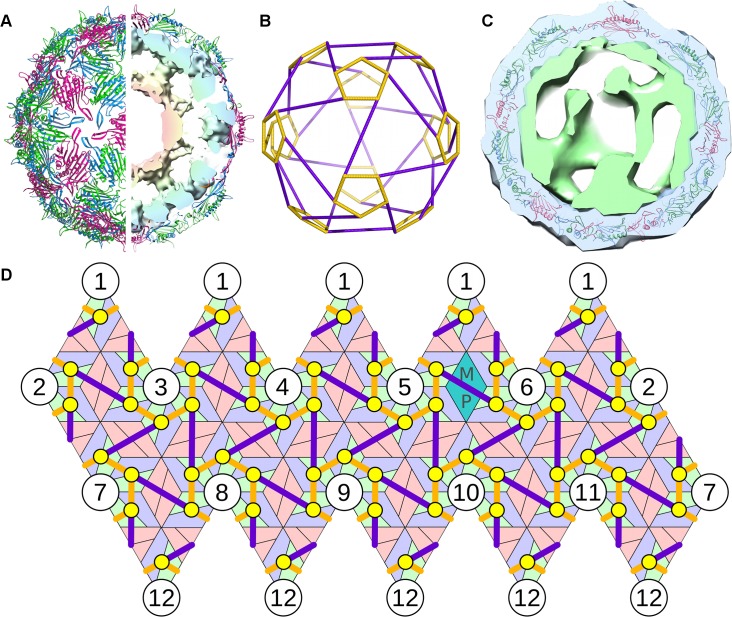
Fig 1. The model system—bacteriophage MS2.
(A) The viral capsid is formed from 60 asymmetric and 29 symmetric copies of the CP dimers, with one MP that takes the place of a symmetric dimer (PDBID 2MS2). The genomic RNA is organized inside the particles in two shells, with the outer shell adopting the shape of a polyhedral cage in icosahedrally-averaged reconstructions. (B) Depiction of the polyhedral cage, showing long (purple) and short (orange) PS-PS RNA connections. (C) Asymmetrically averaged tomogram of bacteriophage MS2 bound to its receptor, the bacterial F-pilus. The portion of the electron density corresponding to the CP shell (and bacterial pilus) is shown in blue; green depicts the density for genomic RNA (and presumably some elements of the MP), which forms the basis for the analysis described in this study. The RNA density forms a shell that is intimately associated with the inside surface of the capsid. (D) A planar representation of protein container and polyhedral RNA organization, showing the relative positions of the 60 polyhedral vertices (PS positions, indicated as yellow circles) in contact with the 60 asymmetric CP dimers.