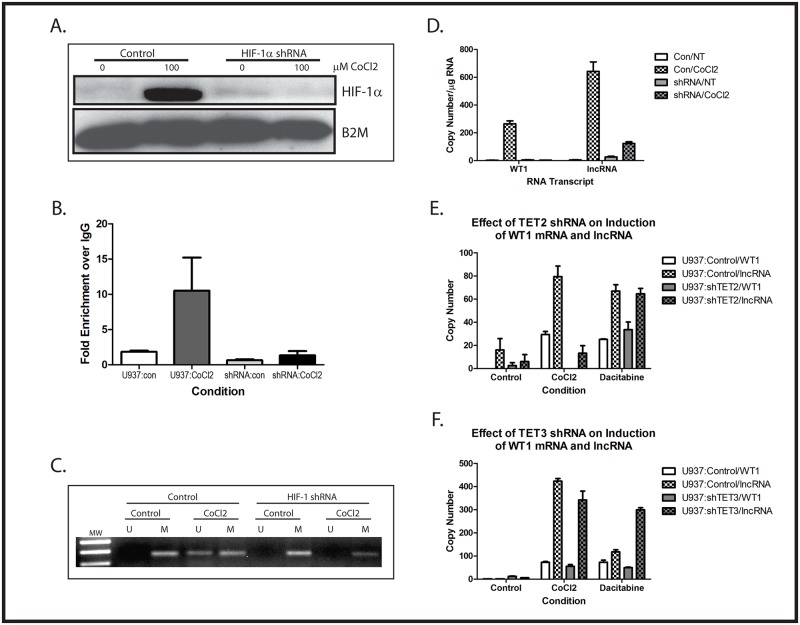
Fig 5. Importance of HIF-1α and TET2/3 in CoCl2-mediated induction of WT1 and WT1 lncRNA.
(A) U937 cells stably transfected with either HIF-1α shRNA or a scramble control were treated with or without 100 μM CoCl2 overnight. Total cellular protein was isolated and analyzed by western blotting with anti-HIF-1α antibody. Stabilization of HIF-1α is seen in CoCl2-treated cells, but this is inhibited by the HIF-1α shRNA. This and the other experiments in this figure were repeated with a second shRNA with similar results. (B) Chromatin immunoprecipitation assays, with quantitative PCR, were performed using antibodies specific for HIF-1α to evaluate binding to the HRE in the WT1 promoter in U937 cells transfected with either the HIF-1α shRNA or a scramble control. Cells were treated with or without 100 μM CoCl2 overnight, and chromatin was isolated, immunoprecipitated with anti-HIF-1α antibody or an isotype control, and analyzed by quantitative PCR using primers surrounding the WT1 HRE. Fold enrichment compared with isotype control was calculated. Data are fold change relative to untreated (control), and error bars show standard error of the mean of triplicate assays. CoCl2 induced a statistically significant (p<0.05) increase in HIF-1α binding to the HRE in control cells, but not in cells stably transfected with HIF-1α shRNA. (C) U937 cells stably transfected with HIF-1α shRNA or a scramble control were treated with or without (control) 100 μM CoCl2 overnight. Genomic DNA was isolated, treated with sodium bisulfite, and the methylation status of the intron 1 CpG island was assessed by methylation-specific PCR. Treatment with CoCl2 caused demethylation of the CpG island in control cells, but not in cells expressing the HIF-1α shRNA. (D) U937 cells transfected with HIF-1α shRNA (shRNA) or a scramble control (Con) were treated with or without (NT = no treatment) 100 μM CoCl2 overnight and analyzed by quantitative RT-PCR using primers specific for WT1 or the WT1 lncRNA. Data are copy number per μg RNA, and error bars show standard error of the mean of triplicate assays. (E) U937 cells stably transfected with TET2 shRNA or scramble control were treated for 48 hours with either 100 μM CoCl2 or 0.3 μM dacitabine and analyzed by quantitative RT-PCR for WT1 and lncRNA expression. Data are shown as copy number per μg RNA, and error bars represent standard error of the mean of triplicate experiments. Statistical analysis using Student’s t test showed significant (p<0.05) increases in both WT1 and lncRNA mediated by dacitabine in both cell lines, but only by CoCl2 in the TET2 knock down cell line. (F) U937 cells stably transfected with TET3 shRNA or scramble control were treated for 48 hours with either 100 μM CoCl2 or 0.3 μM dacitabine and analyzed by quantitative RT-PCR for WT1 and lncRNA expression. Data are shown as copy number per μg RNA, and error bars represent standard error of the mean of triplicate experiments. Statistical analysis using Student’s t test showed significant (p<0.05) increases in both WT1 and lncRNA mediated by both CoCl2 and dacitabine. All experiments were repeated a minimum of 3 times with similar results.