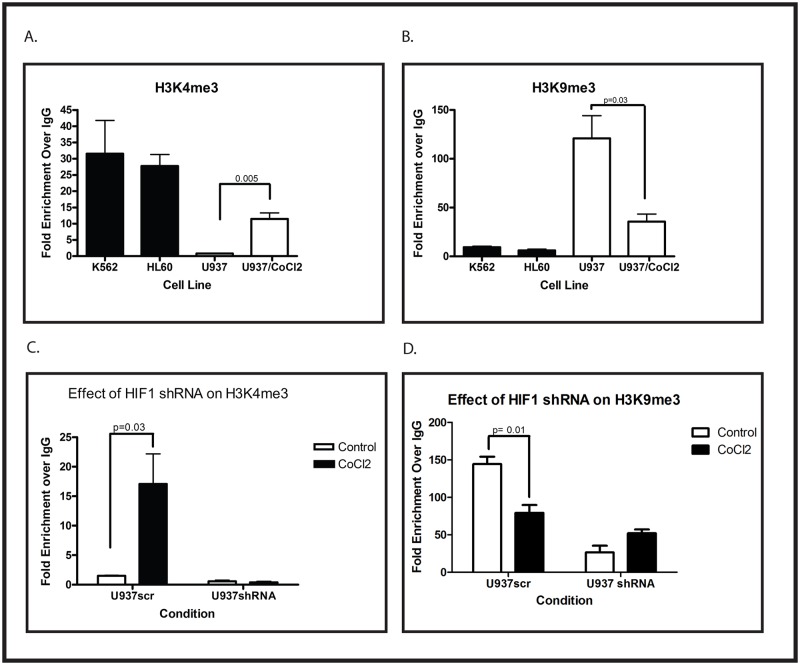
Fig 6. CoCl2-mediated changes in histone methylation around the WT1 transcription start site.
(A and B) Chromatin immunoprecipitation assays, with quantitative PCR, were performed using antibodies specific for H3K4me3 (A) or H3K9me3 (B) on material isolated from K562 cells, HL60 cells, or U937 cells with or without 100 μM CoCl2 treatment for 48 hours. Error bars show standard error of the mean for triplicate assays. The changes induced by CoCl2 were confirmed to be statistically significant (p values shown) by Student’s t test. (C and D) U937 cells stably transfected with an shRNA specific for HIF-1α or a scramble control (scr) were treated with or without 100 μM CoCl2 for 48 hours and analyzed by chromatin immunoprecipitation, with quantitative PCR, using antibodies specific for H3K4me3 (C) or H3K9me3 (D). Statistically significant differences (Student’s t test p value shown) were seen in control (scr) cells but not in cells expressing the shRNA. Error bars show standard error of the mean for triplicate assays. All experiments were repeated at least 3 times with similar results.