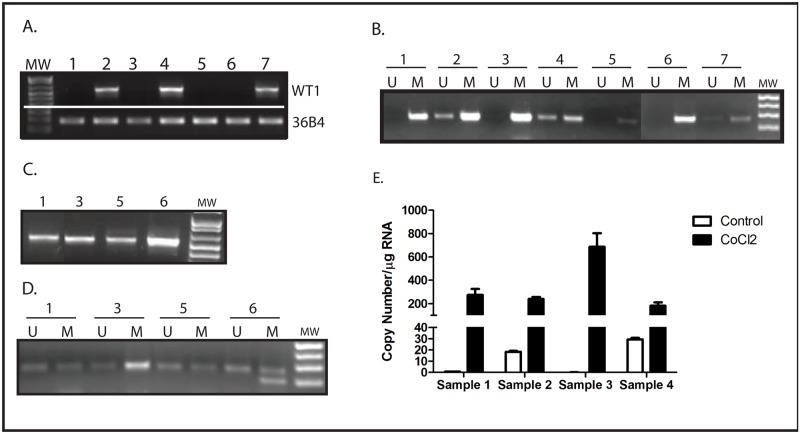
Fig 8. CoCl2 induces CpG island demethylation and WT1 expression in primary AML samples.
(A) Expression of WT1 in 7 independent primary AML samples was evaluated by RT-PCR. The ribosomal RNA 36B4 was used as a loading control. (B) Genomic DNA from the same AML samples was isolated, treated with sodium bisulfite, and evaluated by methylation-specific PCR using primers specific for WT1 intron 1. Unmethylated DNA (U) was only detected in the samples with WT1 expression, and methylated DNA (M) was detected in the samples without WT1. (C) The 4 WT1-negative samples were cultured for 48 hours with 100 μM CoCl2 and WT1 expression analyzed by RT-PCR. (D) Genomic DNA was isolated from the 4 WT1-negative samples after treatment with 100 μM CoCl2. DNA was treated with sodium bisulfite and the methylation status (unmethylated = U, methylated = M) of the intron 1 CpG island was determined by methylation-specific PCR. (E) Expression of WT1 lncRNA was analyzed by quantitative RT-PCR in 4 of the primary AML samples before and after treatment with 100 μM CoCl2. Bars show copy number per μg RNA, and error bars show standard error of the mean for triplicate assays.