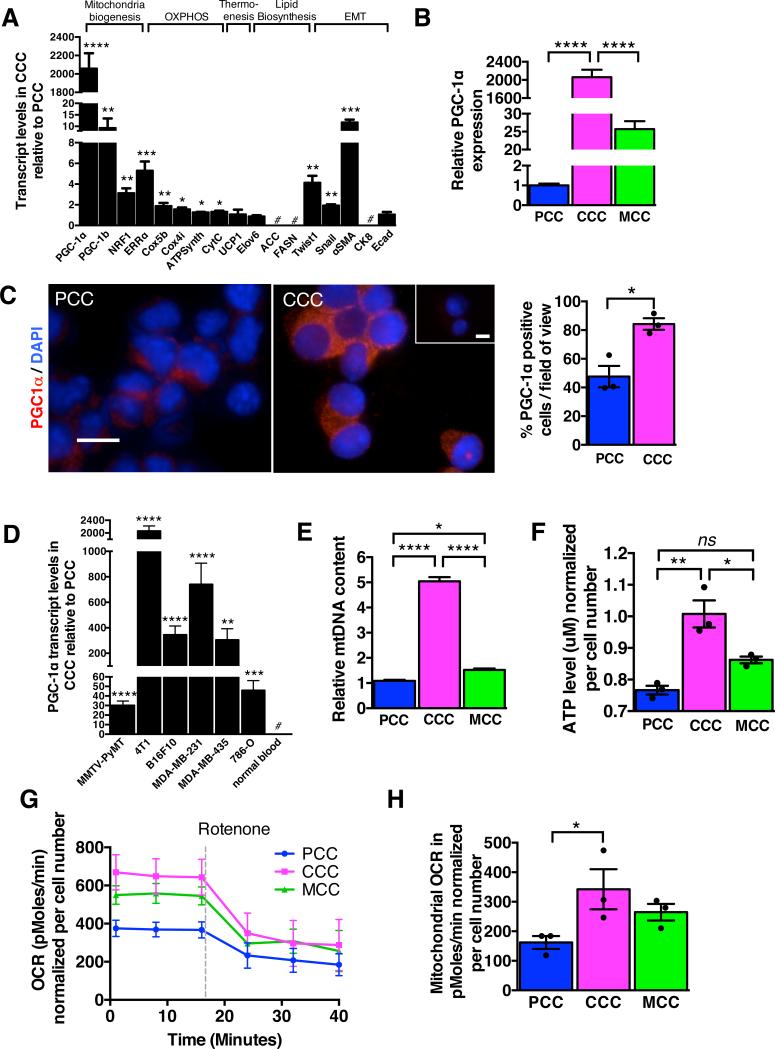
Figure 2. CCC display increased oxygen consumption rate associated with PGC-1α expression and mitochondria biogenesis.
A. Quantitative PCR analyses of relative expression of indicated genes in CCC normalized to PCC. Genes were grouped based on known association with mitochondria biogenesis, oxidative phosphorylation, thermogenesis, lipid biosynthesis, and epithelial to mesenchymal transition (EMT). #: no transcript was detected (n=5 RNA samples from 5 mice, unpaired two-tailed Student's t-test). B. Relative PGC-1α expression by quantitative PCR analysis in CCC compared to PCC in mice with 4T1 orthotopic tumors (n=5 RNA samples from 5 mice, one-way ANOVA). C. Immunostaining for PGC-1α of cytospin of PCC and CCC and quantification of relative percent of PGC-1α positive cells. Scale bars: 10 μm. Nuclear staining (DAPI, blue) (n=3 average percent positive cells from 3 mice, unpaired two-tailed Student's t-test). D. Relative PGC-1α expression by quantitative PCR analysis in CCC compared to PCC in indicated orthotopic tumor models. # (normal blood): no PGC-1α expression detected (n=5 RNA samples from 5 mice, unpaired two-tailed Student's t-test). E. mitochondrial DNA (mtDNA) content (n=5 DNA samples from 5 mice, one-way ANOVA), F. intracellular ATP levels (n=3 lysates from 3 mice, one-way ANOVA), G. oxygen consumption rate (OCR), and H. mitochondrial OCR (delta OCR pre and post rotenone treatment) in PCC, CCC and MCC from 4T1 orthotopic tumor model (n=3 wells of cells from 3 mice, one-way ANOVA). Data is represented as mean +/− SEM. ns: not significant. * p<0.05, ** p<0.01, ** p< 0.001, ****p < 0.0001.