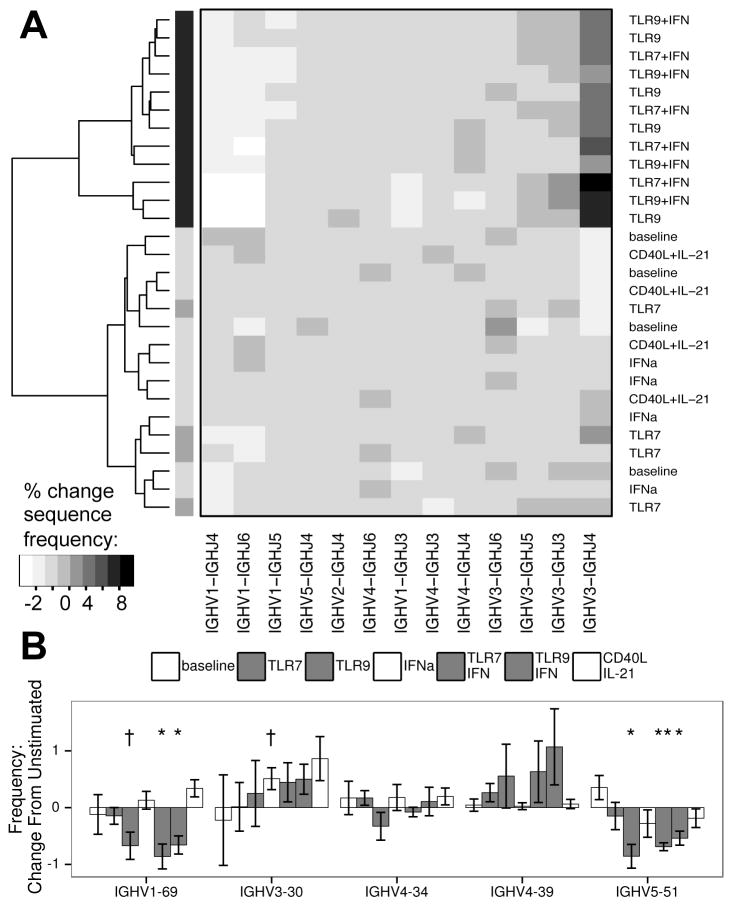
Figure 4. Deep Sequencing of the IgH locus reveals clustering of TLR stimulated cells based on V and J usage.
A: Unbiased clustering of IgHV and IgHJ pairing frequencies of the mutated sequence subset, showing grouping of TLR9 and TLR/IFN (in black) stimulated samples from four donors in contrast to other cultures (dendrogram shading added for ease of viewing). To control for inter-donor differences and nonspecific changes due to culture, data were normalized based on unstimulated samples prior to clustering. B: Changes in frequency of IgHV genes of specific interest (see results section for description). A: Algorithm: complete clustering based on Euclidean distances for IgHV-IgHJ pairings, filtered on pairings with at least 1% change in frequency between any two samples. B: bars represent mean, error bars represent SEM. Columns appear in order of legend. † = p < 0.1; * = p < 0.05; ** = p < 0.01; A–B: data represent four independent donors, split across two independent sequencing runs.