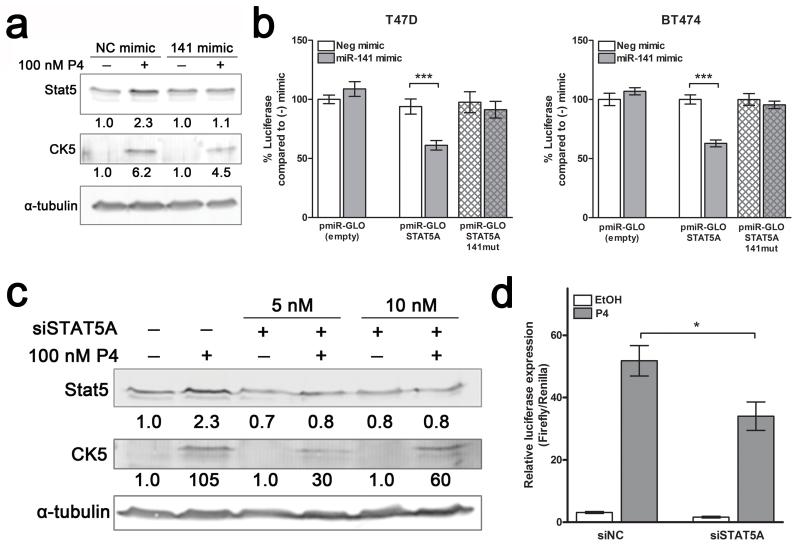
Figure 5.
Stat5a is a direct target of miR-141 and contributes to the P4 expansion of CK5+ cells. (a) P4 increases Stat5 expression, which is partially blocked by miR-141. Western blot analysis of Stat5 and CK5 protein expression in T47D cells treated with EtOH (−) or 100 nM P4 (+), and with negative control mimic (NC) or miR-141 mimic. Fold change of Stat5 and CK5 compared to EtOH/NC mimic is indicated; quantification is normalized to α-tubulin (loading control). (b) Stat5a is directly targeted by miR-141. The STAT5A 3′-UTR was cloned downstream of luciferase in the pMIR-GLO vector and the predicted binding site was mutagenized to abolish miR-141 binding ability. The construct (solid bars) or its mutagenized counterpart (patterned bars) was transfected into T47D or BT474 cells with either 50 nM negative control or miR-141 mimic and luciferase activity measured after 48 h. Data represents relative luciferase activity normalized to constitutively active Renilla contained on the same vector. Experiments were repeated twice. Bars are mean +/− SEM; *** P<0.0001. (c) siSTAT5A blocks P4-mediated upregulation of Stat5. Western blot of Stat5 and CK5 protein expression of T47D cells treated with EtOH or 100 nM P4, with or without 5 or 10 nM siSTAT5A. α-tubulin was used as loading control. Fold changes in Stat5 and CK5 relative to vehicle with negative control siRNA are indicated. (d) siSTAT5A inhibits P4 induction of CK5+. T47D cells stably expressing a luciferase reporter driven by the CK5 gene (KRT5) promoter were plated at 10,000 cells per well in 96-well plates. After 24 h, cells were transfected with SV40-Renilla vector and 5 nM negative control (siNC) or On-Target pool siSTAT5A using dual transfection reagent. Cells were treated for an additional 24 h with vehicle (EtOH) or 100 nM P4, and luciferase activity measured. Renilla was used to normalize luciferase data for transfection efficiency. Bars are mean +/− SEM. *P<0.05.