Abstract
At present there is no well structured database available for the venomous snakes and venom composition of snakes in the world although venom has immense importance in biomedical research. Searching for a specific venom component from NCBI, PDB or public databases is troublesome, because they contain huge amount of data entries. Therefore, we created a database named “ISOB” which is a web accessible unique secondary database that represents the first online available bioinformatics resource showing venom composition of snakes. This database provides a comprehensive overview of seventy-eight indigenous snake species covering description of snakes supplemented with structural information of the relevant individual available venom proteins. We strongly believe that this database will contribute significantly in the field of bioinformatics, environmental research, proteomics, drug development and rationale drug designing.
Availability
The database is freely available at http://www.snakebd.com/
Keywords: Snake venom, Database, Indigenous snakes, Bangladesh, 3D structure
Background
Snakes are a group of amazing reptiles which have fascinated human being since beginning of mankind. There are about 3000 different snake species described on earth, of these, only about 350 are regarded as poisonous and dangerous for humans. Although snakes are an important parameter of our ecosystem and have tremendous importance in biopharmaceutical research, that been ignored for proper recording possibly due to the danger and panic of handling of this mysterious reptiles. In snakes, venom is very important which is produced and stored in highly specialized venom glands and used mainly for defense or to immobilize prey [1]. Each of these proteins and peptides has profound effect on various physiological systems of victim [2, 3]. More than a century of research has shown that snake venom is a rich resource for pharmacologically active proteins and peptides [4] and good natural source of drug lead compounds. A number of drugs are currently available in the market derived from snake venom proteins and hundreds are in the different stages of developmental pipeline [5]. For example, Captopril® is the most extensively used drug to control blood pressure and cardiac diseases, was developed from one of the venom components of pit viper snake (Bothrops jararaca). Scientists and clinicians in the pharmaceutical development field utilize the molecules of snake venom as therapeutic agents for several pathologies such as hypertension, cancer, thrombosis [6]. In spite of such huge importance there is no separate database available for snake venom proteins. ‘The Reptile Database’ [7] is the only snake related database which merely covers the descriptive and taxonomical aspect of different snake species of the world. Nevertheless, the venom components which have great pharmacological interest are not included in the database. Searching of a specific venom component with particular interest and specific physiological function from NCBI, PDB or public databases are time consuming and troublesome because these databases contain huge amount of data entries. These facts forced to develop a separate database for the snake venoms proteins.
Bangladesh is a country in South Asia, located on the fertile ‘Bengal delta’ with rich flora and fauna. Despite of having great promise, snakes received no appropriate attention from the planners and professionals of this country. Snake population in Bangladesh is at high risk due to overwhelming population growth and reclamation of forest and wildlife habitat to mitigate the increasing land requirement for accommodation and food. Only a few documentations are present for snakes in Bangladesh. According to the report published by IUCN in 2000 there are about 98 species of snakes in Bangladesh, although over a dozen of these have not been reported during the last few decades. Therefore, there is an imperative need to have a database covering the venom components of the snake along with their structural and functional information. These facts motivated to develop a secondary database titled “ISOB: A Database of Indigenous Snake Species of Bangladesh with respective known venom composition” with detailed information available so far for seventy-eight indigenous snake species covering description of snakes along with the information of available regarding the venom proteins. It has a set of search tools that permit the users to extract data and carry out specific queries.
Database description
The aim of ISOB is to offer a unique assortment of data extracted from both public databases and published reports which help to retrieval of information and the analysis of stored data. This database supplies hyperlinks to the corresponding entries of the related external databases. The ISOB database contains information of 78 species of snakes exhibiting relevant information about them. Since January 2013, it contained a total of 405 of snake toxins.
Data collection resources
Data of snakes found in Bangladesh were collected from various resources such as list of Dr. Ali Reza Khan [8], IUCN Red List [9] and Reptile Database. Research papers and Case reports from Bangladesh, India, Pakistan, Bhutan, Nepal, and Sri Lanka were retrieved, as were significant papers from other Asian countries and also from different available informative websites. Moreover, the entries of venom components have been compiled from different public databases such as PubMed [10], Genbank [11], Swissprot and TREMBL [12] and PDB [13]. The number of entries extracted from several sources is given in Table 1 (See Supplementary material).
Database design
ISOB is developed using PHP (Hypertext preprocessor) as server-side scripting language and embedded in HTML. MySQL is applied as back-end database management system [14], because of its capability to maintain very large databases. It is a user-friendly web-based application which connects to the database of the wide-ranging indigenous snake species of Bangladesh. The web application has been constructed with a simple and advanced search options. In simple search approach, one can search for a snake for its general information along with their venom components including their three-dimensional structures (either from PDB or predicted by molecular modeling) by directly using its family, genus, species or scientific name while the advanced search option has been built to search for venoms of any particular class those are present in available venomous snake species of Bangladesh. For selection of multiple sequences from each class a checkbox has been created beside each of the sequence. Using these selected sequences can be exported in a text file as FASTA format which will ease various bioinformatics analysis such as multiple sequence alignment, similar protein search using BLAST etc. The grouping of snake toxins provides a basis for extending and clarifying the existing structural and functional classification. Among all these venom components, 102 numbers of three dimensional structures were retrieved from protein databank and remaining 253 number of new unsolved protein structures were predicted using molecular modeling method SWISS-MODEL [15]. SWISS-MODEL is an entirely automated protein structure homology-modeling server, accessible through the ExPASy web server, or from the DeepView (Swiss Pdb-Viewer) program. The use of this server is to make Protein Modeling accessible to all biochemists and molecular biologists worldwide [16]. The models were created with the help of Rasmol software by using the PDB template generated from SWISSMODEL. This will ease the bioinformatics analyses for identification of sequence patterns associated with specific structural or functional properties of snake toxins. The internet browser accessible interface ISOB database is shown in Figure 1.
Figure 1.
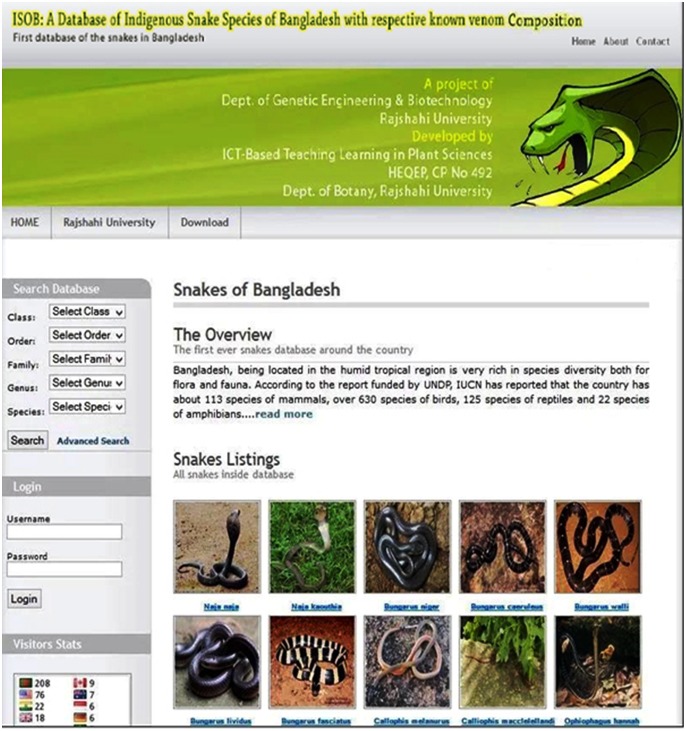
Home page of the database (http://www. snakebd. com)
Features of the ISOB database records
The ISOB database exists in as an open access, public domain secondary web database. This database acts as a complete web source for seventy-eight indigenous snake species of Bangladesh along with the information regarding their venom components. Out of 78, 31 snake species are venomous, 42 are mildly venomous and 5 are non-venomous. Among these 31 venomous snake species of Bangladesh, 405 venom components from 13 species are available to date. All the venom protein sequence information was systematically arranged in this database. Each of the records in the ISOB database has a unique documentation of each snake species encloses information about (1) Images with their sources (2) Common name (3) Local name (4) Scientific Name (5) Venom presence (6) Scientific Classification (7) Synonym (8) Distribution (9) Size (10) Structural features (11) Habitat (12) Diet (13) Reproduction (14) Behavior (15) Venom characteristics (16) Key characteristics for identification and (17) Venom (The proteins can be queried by suggesting the accession number). This venom section further has sub sections and is designed by taking eight important parameters into consideration viz. (i) Image (ii) Venom title (iii) Class (iv) Accession (v) Reference (vi) Protein sequence (vii) Structural description and (viii) Related snake. Snake toxin proteins are grouped into two main superfamilies: enzymatic and nonenzymatic proteins [17]. The non enzymatic venom components includes Three-finger toxins, C-type lectin-related proteins, Kunitz type-serine proteinase inhibitors, Sarafotoxins, Cysteine-rich secretory protein (CRISP), C-type lectins, Disintegrins, Waprins, Veficolins, Vespryns, Helveprins and Myotoxins. On the other hand enzymatic proteins in venom include Phospholipase A2 enzymes, Proteinases (Serine proteinases and Metalloproteinases), Nucleotidases, Amino acid oxidase and Acetylcholinesterase. So far, different classed venom components present in different venomous snake species from Bangladesh are integrated in this database. A representative entry of the entire data record is given as a flowchart (Figure 2).
Figure 2.
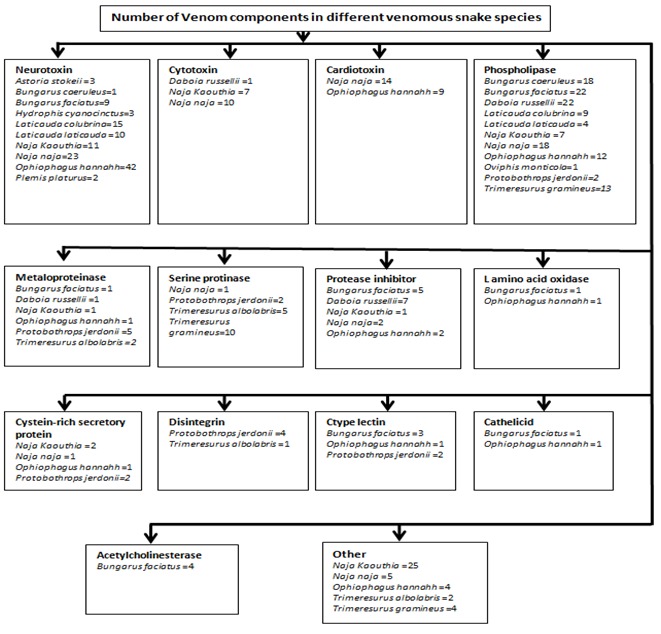
Flow chart
During this investigation, several errors were revealed in data from public sources and discrepancies of information supplied for the same toxins between different sources. However, In ISOB venom gallery, each of the venom has a unique accession number of the sequences of the snake database entries have that have corresponding entries in public databases such as Genbank, Swissprot and PDB and provides hyperlinks to the corresponding entries in these source databases. A representative entry of the overall data record is given in Figure 3. Protein table consists of the protein sequence with snake wise specific classes are unique feature of this database (Figure 4). Snake venom contains wide ranges of several structurally distinct families of peptides those have miscellaneous functions. As, snake bite is a burning public health issue and second most common cause of un-natural fatality in Bangladesh, ISOB included information regarding snake bite treatment in local language. Apart from the technical information, the database also gives the up to date state of the availability status of snakes in Bangladesh.
Figure 3.

An example of snake database overview (a) Total data record of specific snake species (b) Retrieved PDB protein structure and (c) Predicted new protein structure.
Figure 4.

An example of advanced search page showing protein sequence of a particular class of protein from different snakes.
Discussion
Venomous snakes are found in the family׳s Colubridae (boomslang, vine snake), Elapidae (cobra, krait, mamba, taipan, tiger snake, coral snake), Hydrophiidae (sea snakes), Viperidae (old world vipers found in Europe, Africa, Asia but not in America or Australia, saw scaled viper, Russell's viper, puff adder, Gaboon viper), and Crotalidae (pit vipers, found in America, Asia and Europe, copperhead, cotton mouth, rattlesnakes [18]. In this current study mainly focused on the snakes found in Bangladesh. These snakes are mostly from the Elapidae,Viperidae, Hydrophiidae and colubridae families. Venom is a complex mixture of proteins and peptides (most of which reveal enzymatic activity), amines, lipids and other components which targets specifically different physiological systems of victim or pray [19]. Due to high activity and high specificity of venom components, they are useful for the study of highly complex biological mechanisms. A wide array of animals including terrestrial: ants, wasps, scorpions, spiders and snakes and marine organisms: Jellyfish, sea anemones, puffer fish, cone snails, sea snakes and octopuses produced venom. Rough estimates of the number of different venom components from individual snake, scorpion or cone snail is in the range of 50–200 different toxins [20] with the growing number of newly characterized toxins, their arrangements will require updating and need better data management. Till now information relating venom-toxin sequences and the features, structure and function of venom-toxins is scattered across multiple sources. Only basic sequence information is found in Primary databases such as GenBank or SWISS-PROT. These public databases contain huge amount of data entries which may be unnecessary for bioinformatics analysis of snake venom components with particular interest. For example, a basic search for all snake venom Phospholipase A2 (svPLA2) in the GenBank and SWISSPROT databases results 55% of irrelevant and redundant sequence entries (including pancreatic PLA2, bacterial PLA2 and PLA2 from other non venomous sources) and required to be filtered out before the analysis. Therefore, a fresh and comprehensive database comprising the description on the snakes supplemented with structural information of the relevant individual venom components available so far will be of special interest for the bioinformatics researchers. As far as our knowledge, only particular venom databases are at present as major sources for the study of venom toxin. The databases include entries collected from different sources, cleaned, organized, analyzed and classified in keeping with their structure–function relationship. The SCORPION database [21] of just about 300 entries of scorpion toxin sequences are annotated and classified along with their structural and functional properties. The Mollusk [22] database comprises over 450 peptides from the cone snail venoms where each entry has a distinctive field to make easy comparison of conotoxin entries. Functionally interpreted entries of svPLA2 toxins are found in the svPLA2 database [23]. However, currently there is no separate database for snake venom components. Therefore, the Database titled ISOB; a secondary database has been developed in the current project with detailed information available so far for 78 available snake species of Bangladesh. In this database, the venom has been grouped in different classes for the easy of searching a particular type of venom component among and across species. Three dimensional structures (3D) of a molecule have enormous implication and importance in Biological Sciences which offer a complementary approach to sitedirected mutagenesis for identification of functional residues in venom toxins [24] aid in structure-function studies and also help in rationale drug designing. Structure determination by NMR is limited only to small peptide molecules whereas macromolecules crystallization is a slow and complex process, which involves optimization of various interdependent physical, chemical and biological parameters. Therefore, the prediction of 3D structures of proteins from primary structures by comparative protein modeling techniques is an attractive alternative for large number of toxins.
Conclusion
A database for the indigenous snakes of Bangladesh is created and for the first time all available information regarding the snake venom proteins have been integrated and arranged under one umbrella. Three dimensional structure of 253 unsolved venom protein structure using homology modeling are also incorporated in the database. Such a huge number of predicted 3D structure will help in studying structure-function relationship, pharmacological action, protein evolution, developing mini protein scaffold and in numerous other bioinformatics analysis. This secondary database is a great initiative and it is strongly believed that scientist throughout the world will be benefited from the database and that will contribute in different bioinformatics investigations. We envision this database as a template for management of molecular data of toxins of various venomous snake species. Thus, this database is specialized for molecular analysis of snake toxins which will serve multidimensional research purposes. This database will serve as a basic guideline for future field based wild-life survey on reptiles of Bangladesh and it will be easier for the field researcher for identification of the available snake species of Bangladesh. It also assists to build up awareness of peoples for primary management and treatment during snake bite.
Future Development
In future, refinement and periodical update of the database will be carried out so as to ensure that users get the latest information. It is also intended to include more sophisticated Bioinformatics tools into the database for facilitating in-depth structure–function studies, characterization of toxin interaction sites and drug discovery.
Supplementary material
Acknowledgments
The authors would like to thank the Higher Education Quality Enhancement Project (HEQEP) for financial assistance (HEQEP CP no. 492) and technical support. The authors would also like to thank Professor R.M. Kini, Department of Biological Sciences, National University of Singapore for his valuable comments and suggestions and the Snake Rescue and Conservation Centre (SRCC) of Darusha, Rajshahi, Bangladesh for a number of valuable information.
Footnotes
Citation:Roly et al, Bioinformation 11 (2): 107-114 (2015)
References
- 1.Roly ZY, et al. Bioinformation. 2014;10:617. doi: 10.6026/97320630010617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Q, et al. J Thromb Haemost. 2005;3:1791. doi: 10.1111/j.1538-7836.2005.01358.x. [DOI] [PubMed] [Google Scholar]
- 3.Markland FS. Toxicon. 1998;36:1749. doi: 10.1016/s0041-0101(98)00126-3. [DOI] [PubMed] [Google Scholar]
- 4.Roly ZY, et al. Bioinformation. 2014;10:281. doi: 10.6026/97320630010281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox JW, Serrano SM. Curr Pharm Des. 2007;13:2927. doi: 10.2174/138161207782023739. [DOI] [PubMed] [Google Scholar]
- 6.Smith CG, Vane JR. FASEB J. 2003;17:788. doi: 10.1096/fj.03-0093life. [DOI] [PubMed] [Google Scholar]
- 7. http://reptile-database.org/
- 8.Khan MAR, et al. Bangladesher banya prani. 1987 [Google Scholar]
- 9.UCN-Bangladesh. 2000 Red Book of Threatened Amphibians and reptiles of Bangladesh. IUCN, The World Conservation Union. [Google Scholar]
- 10.Wheeler DL, et al. Nucleic Acids Res. 2001;29:11. [Google Scholar]
- 11.Benson DA, et al. Nucleic Acids Res. 2000;28:15. [Google Scholar]
- 12.Bairoch A, Apweiler R. Nucleic Acids Res. 2000;28:45. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman HM, et al. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alekseyenko AV, Lee CJ. Bioinformatics. 2007;23:1386. doi: 10.1093/bioinformatics/btl647. [DOI] [PubMed] [Google Scholar]
- 15. http://swissmodel.expasy.org/
- 16.Kiefer F, et al. Nucleic Acids Res. 2009;37:D387. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang TS, et al. FEBS J. 2011;278:4544. doi: 10.1111/j.1742-4658.2011.08115.x. [DOI] [PubMed] [Google Scholar]
- 18.Karalliedde L. Br J Anaesth. 1995;74:319. doi: 10.1093/bja/74.3.319. [DOI] [PubMed] [Google Scholar]
- 19.Church JE, Hodgson WC. Toxicon. 2002;40:1083. doi: 10.1016/s0041-0101(02)00126-5. [DOI] [PubMed] [Google Scholar]
- 20.Lourenco WR. Biogiographica. 1994;70:155. [Google Scholar]
- 21.Srinivasan KN, et al. Toxicon. 2002;40:23. doi: 10.1016/s0041-0101(01)00182-9. [DOI] [PubMed] [Google Scholar]
- 22. http:// sdmc. krdl. org. sg: 8080/ MOLLUSK/
- 23. http://sdmc.lit.org.sg/Templar/DB/snaketoxin_PLA2/i ndex.html.
- 24.Renisio JG, et al. Proteins. 2000;38:70. doi: 10.1002/(sici)1097-0134(20000101)38:1<70::aid-prot8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


