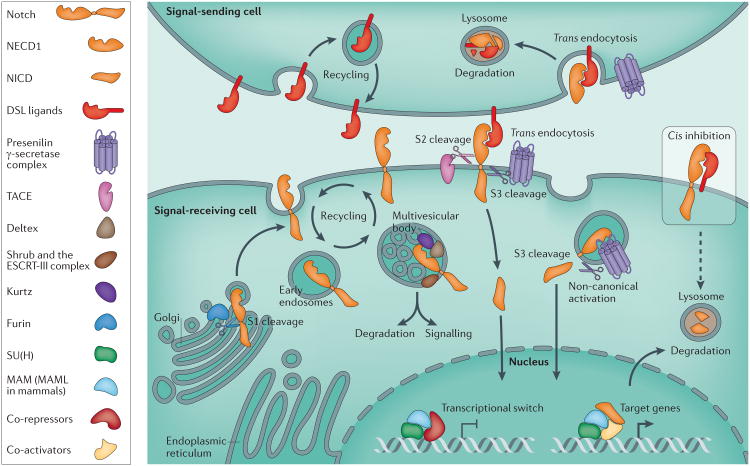
Figure 1. Summary of the main features of Notch signalling.
Three proteolytic cleavage steps are required for canonical Notch receptor signalling. The first proteolytic cleavage step (S1 cleavage) is mediated by Furin, occurs in the trans-Golgi and produces a heterodimer composed of a ligand-binding Notch extracellular domain (NECD) and a single-pass transmembrane signalling domain referred to as the Notch intracellular domain (NICD). The functional importance of this cleavage is still somewhat unclear130,131. The association of NECD and the transmembrane portion of the receptor heterodimer is dependent on non-covalent interactions. Pathway activation occurs when the NECD binds to Delta– Serrate–LAG2 (DSL) ligands that are expressed on the membrane of neighbouring cells. This trans interaction results in the second proteolytic event (S2 cleavage) of the Notch receptor, which clears most of the NECD from the outer portion of the membrane, a process mediated by the TACE (also known as ADAM17) metalloproteinase. The NECD is subsequently released and internalized through endocytosis by the ligand-expressing cell, where it undergoes lysosomal degradation. Subsequently, γ-secretase cleaves the tethered receptor near the inner leaflet of the membrane (S3 cleavage) in the Notch-expressing cell, producing the transcriptionally active NICD, which translocates to the nucleus through a poorly understood process. In the nucleus, the NICD interacts with Drosophila melanogaster Suppressor of Hairless (SU(H)) and the transcriptional co-activator, Mastermind (MAM) — the mammalian orthologues of which are CBF1–SU(H)–LAG1 (CSL) and Mastermind-like (MAML) proteins, respectively — thereby inducing transcription of target genes, by converting CSL into a transcription activator through the exchange of co-repressors for co-activators. Many Notch target genes encode transcriptional regulators, which influence cell-fate decisions through the regulation of basic helix–loop–helix hairy and enhancer of split (HES) proteins: Hairy and Enhancer of split (E(SPL)) in D. melanogaster and their mammalian orthologues HES1 and HES5. The HES proteins subsequently regulate the expression of genes involved in Notch-dependent cell-fate determination, such as apoptosis, proliferation or differentiation. By contrast, expression of ligands and the Notch receptor on the same cells results in cis inhibition of Notch signals and receptor degradation. Recycling the receptor through the endocytic pathway has been shown to be important for receptor and ligand maturation, non-canonical signalling and degradation (Box 2). Notch activity is regulated by ubiquitylation of nuclear NICD by the E3 ubiquitin ligases, SEL-10 in Caenorhabditis elegans and Suppressor of Deltex (SU(DX)) in D. melanogaster, leading to NICD degradation, thus allowing the cell to become ligand-competent once again. The ubiquitylation status of the receptor (by Kurtz, Deltex and Shrub) in the multi-vesicular bodies can also determine whether Notch continues to signal or undergoes proteasomal degradation. Additionally, signal attenuation is achieved through lysosomal degradation of the NICD.