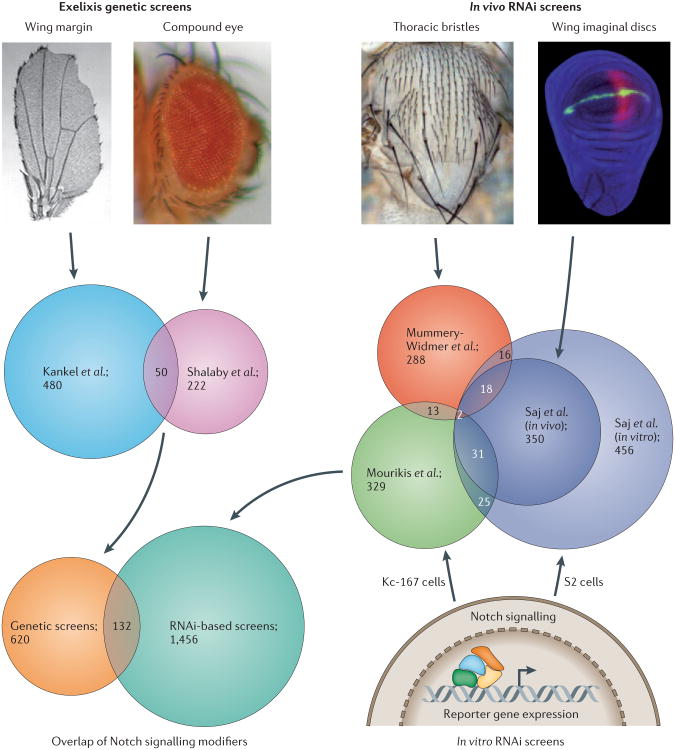
Figure 2. Overlap of Notch genetic modifiers from different genome-wide screens in Drosophila melanogaster.
The first screen to take advantage of the Exelixis collection of insertional mutations was carried out by Kankel et al.36, and the authors screened for modifiers of a Drosophila melanogaster wing margin phenotype that resulted from ectopic expression of dominant negative version of Mastermind (MAM). The second screen to use the Exelixis collection was carried out by Shalaby et al.37 and isolated modifiers of the cell-fate alterations in the D. melanogaster eye that are caused by overexpression of the Notch ligand Delta. Among the genes identified from these two independent genetic screens, 50 genes were observed in both studies. Genome-wide RNAi screens were carried out by several different groups using in vivo developmental phenotypes associated with the Notch pathway or in vitro cell-based Notch-signalling-dependent reporter assays. Mummery-Widmer et al.39 used external sensory organ development during thoracic bristle formation as a phenotypic parameter to isolate genes that affect Notch signalling, whereas Saj et al.38 carried out a genome-wide screen in S2 cells for regulators of Notch activity. Nearly half of the genes identified in the primary S2-cell-based screen were subsequently in vivo validated in developing wing imaginal discs. Mourikis et al.51 measured the transcriptional response of a luciferase-reporter (m3-luc) in Kc-167 cells to identify modifiers of Notch activity. However, the overlap of genes identified in all of the in vivo and in vitro RNAi screens is small (consisting of only two genes). From these screens, a total of 2,208 genes (14.25% of 15,494 genes based on FlyBase release 5.43) were found to affect Notch signalling (Supplementary information S4 (table)), 132 of which were identified in both Exelixis-based and RNAi-based screens. The ‘Wing margin’ image is reproduced, with permission, from REF. 36 © (2007) Genetics Society of America. The ‘Thoracic bristles’ image is reproduced, with permission, from REF. 132. The ‘Wing imaginal discs’ image is reproduced from REF. 38 © (2010) Cell Press.