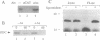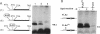Abstract
We provide here an example of a mammalian cellular gene expressed by frame-shifting. Conventional reading of the sequence of ornithine decarboxylase-antizyme mRNA (a protein that modulates the rate of ornithine decarboxylase degradation) results in premature termination at an in-frame termination codon (stop-1), located shortly after the initiation codon. By translating, in vitro in reticulocyte lysate, antizyme mRNA with a full coding capacity and various mutants derived from it, we demonstrate that antizyme expression requires that ribosomes shift from the first open reading frame (termed ORF0) to a second +1 open reading frame (ORF1). Our studies show that this frame-shifting, which occurs at maximal efficiency of approximately 20%, is stimulated by polyamines and requires the functional integrity of the stop codon (stop-1) of ORF0. By introducing in-frame deletions, we have shown that an 87-nt segment surrounding stop-1 enhances frame-shifting efficiency, whereas the 6 nt located just upstream to stop-1 are absolutely essential for this process. Because this segment does not contain sequences that were previously characterized as shifty segments, our results suggest that another mechanism of frame-shifting is involved in mediating antizyme expression.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhar I., Engelberg-Kulka H. A procedure for amino acid sequencing in internal regions of proteins. Gene. 1991 Jul 15;103(1):79–82. doi: 10.1016/0378-1119(91)90394-q. [DOI] [PubMed] [Google Scholar]
- Benhar I., Engelberg-Kulka H. Frameshifting in the expression of the E. coli trpR gene occurs by the bypassing of a segment of its coding sequence. Cell. 1993 Jan 15;72(1):121–130. doi: 10.1016/0092-8674(93)90056-v. [DOI] [PubMed] [Google Scholar]
- Benhar I., Miller C., Engelberg-Kulka H. Frameshifting in the expression of the Escherichia coli trpR gene. Mol Microbiol. 1992 Oct;6(19):2777–2784. doi: 10.1111/j.1365-2958.1992.tb01457.x. [DOI] [PubMed] [Google Scholar]
- Bercovich Z., Rosenberg-Hasson Y., Ciechanover A., Kahana C. Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. J Biol Chem. 1989 Sep 25;264(27):15949–15952. [PubMed] [Google Scholar]
- Blinkowa A. L., Walker J. R. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990 Apr 11;18(7):1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986 Jul 17;322(6076):273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Translational frameshifting: where will it stop? Cell. 1987 Jul 3;50(1):1–2. doi: 10.1016/0092-8674(87)90652-0. [DOI] [PubMed] [Google Scholar]
- Craigen W. J., Cook R. G., Tate W. P., Caskey C. T. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Zhao H., Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993 Jul 16;74(1):93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower A. M., McHenry C. S. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990 May;87(10):3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong W. F., Heller J. S., Canellakis E. S. The appearance of an ornithine decarboxylase inhibitory protein upon the addition of putrescine to cell cultures. Biochim Biophys Acta. 1976 Apr 23;428(2):456–465. doi: 10.1016/0304-4165(76)90054-4. [DOI] [PubMed] [Google Scholar]
- Garcia A., van Duin J., Pleij C. W. Differential response to frameshift signals in eukaryotic and prokaryotic translational systems. Nucleic Acids Res. 1993 Feb 11;21(3):401–406. doi: 10.1093/nar/21.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland R. F., Weiss R. B., Atkins J. F. Recoding: reprogrammed genetic decoding. Science. 1992 Sep 18;257(5077):1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Ghoda L., Sidney D., Macrae M., Coffino P. Structural elements of ornithine decarboxylase required for intracellular degradation and polyamine-dependent regulation. Mol Cell Biol. 1992 May;12(5):2178–2185. doi: 10.1128/mcb.12.5.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heby O., Persson L. Molecular genetics of polyamine synthesis in eukaryotic cells. Trends Biochem Sci. 1990 Apr;15(4):153–158. doi: 10.1016/0968-0004(90)90216-x. [DOI] [PubMed] [Google Scholar]
- Heller J. S., Fong W. F., Canellakis E. S. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm I., Persson L., Stjernborg L., Thorsson L., Heby O. Feedback control of ornithine decarboxylase expression by polyamines. Analysis of ornithine decarboxylase mRNA distribution in polysome profiles and of translation of this mRNA in vitro. Biochem J. 1989 Mar 1;258(2):343–350. doi: 10.1042/bj2580343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. M., Ao S. Z., Casjens S., Orlandi R., Zeikus R., Weiss R., Winge D., Fang M. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988 Feb 26;239(4843):1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- Jacks T. Translational suppression in gene expression in retroviruses and retrotransposons. Curr Top Microbiol Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- Kahana C., Nathans D. Translational regulation of mammalian ornithine decarboxylase by polyamines. J Biol Chem. 1985 Dec 15;260(29):15390–15393. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C. G. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- Li X., Coffino P. Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol Cell Biol. 1993 Apr;13(4):2377–2383. doi: 10.1128/mcb.13.4.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol Cell Biol. 1992 Aug;12(8):3556–3562. doi: 10.1128/mcb.12.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S., Kanamoto R., Murakami Y., Hayashi S. Monoclonal antibody studies on the properties and regulation of murine ornithine decarboxylase antizymes. J Biochem. 1990 Jan;107(1):87–91. doi: 10.1093/oxfordjournals.jbchem.a123018. [DOI] [PubMed] [Google Scholar]
- Matsufuji S., Miyazaki Y., Kanamoto R., Kameji T., Murakami Y., Baby T. G., Fujita K., Ohno T., Hayashi S. Analyses of ornithine decarboxylase antizyme mRNA with a cDNA cloned from rat liver. J Biochem. 1990 Sep;108(3):365–371. doi: 10.1093/oxfordjournals.jbchem.a123207. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y., Matsufuji S., Hayashi S. Cloning and characterization of a rat gene encoding ornithine decarboxylase antizyme. Gene. 1992 Apr 15;113(2):191–197. doi: 10.1016/0378-1119(92)90395-6. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Hayashi S. Role of antizyme in degradation of ornithine decarboxylase in HTC cells. Biochem J. 1985 Mar 15;226(3):893–896. doi: 10.1042/bj2260893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992 Dec 10;360(6404):597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji S., Miyazaki Y., Hayashi S. Destabilization of ornithine decarboxylase by transfected antizyme gene expression in hepatoma tissue culture cells. J Biol Chem. 1992 Jul 5;267(19):13138–13141. [PubMed] [Google Scholar]
- Murakami Y., Tanaka K., Matsufuji S., Miyazaki Y., Hayashi S. Antizyme, a protein induced by polyamines, accelerates the degradation of ornithine decarboxylase in Chinese-hamster ovary-cell extracts. Biochem J. 1992 May 1;283(Pt 3):661–664. doi: 10.1042/bj2830661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. Errors and alternatives in reading the universal genetic code. Microbiol Rev. 1989 Sep;53(3):273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson L., Holm I., Heby O. Translational regulation of ornithine decarboxylase by polyamines. FEBS Lett. 1986 Sep 15;205(2):175–178. doi: 10.1016/0014-5793(86)80892-4. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Nagasawa H., Ohtsubo E. Identification of the site of translational frameshifting required for production of the transposase encoded by insertion sequence IS 1. Mol Gen Genet. 1992 Nov;235(2-3):317–324. doi: 10.1007/BF00279376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. DNA sequences required for translational frameshifting in production of the transposase encoded by IS1. Mol Gen Genet. 1992 Nov;235(2-3):325–332. doi: 10.1007/BF00279377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. Retroviruses. Science. 1988 Jun 10;240(4858):1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Huang W. M., Dunn D. M. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990 Jul 13;62(1):117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]