Figure 3.
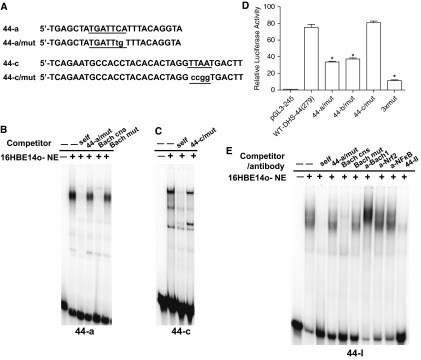
BTB and CNC homology 1, basic leucine zipper transcription factor (Bach1) binds to an antioxidant response element within DHS-44(279) in vitro. (A) Scanning mutagenesis identified the critical nucleotides for transcription factor binding at 44-a and 44-c. (B, C, and E) EMSA experiments with 32P-labeled 44-a (B), 44-c (C), or 44-I (E) probes and 16HBE14o- nuclear extract. The DNA–protein complex generated with each probe was competed by 50× excess of the same unlabeled oligonucleotide but not by mutant versions (44-a/mut and 44-c/mut) (A, Table E3). The complex formed with the 44-a probe was also competed by an unlabeled Bach1 consensus oligonucleotide (39, 40) (Table E3) but not by a mutant version (B). (D) Mutagenesis of DHS-44(279) decreases its enhancer activity, confirming critical transcription factor binding sites. pGL3 constructs and transient reporter gene assays in 16HBE14o- cells as described in Figure 1. Mutant plasmids (44-a/mut and 44-c/mut are shown in A; 44-b/mut is shown in Fig. 2A) were compared with wild-type DHS-44(279). Data show luciferase activities relative to the CFTR basal promoter vector (= 1). Error bars represent SEM (n = 6). *P < 0.05 using unpaired t tests. (E) Binding of Bach1 to 44-I in vitro. The DNA–protein complexes formed with 32P-labeled 44-I probe were competed by 50× excess of unlabeled 44-I and Bach1 consensus probe but not by mutant versions (Table E3). A supershift is seen on incubation of the DNA–protein complex with an antibody specific for Bach1 (NF-κB or nuclear factor, erythroid 2-like 2, isotype controls).
