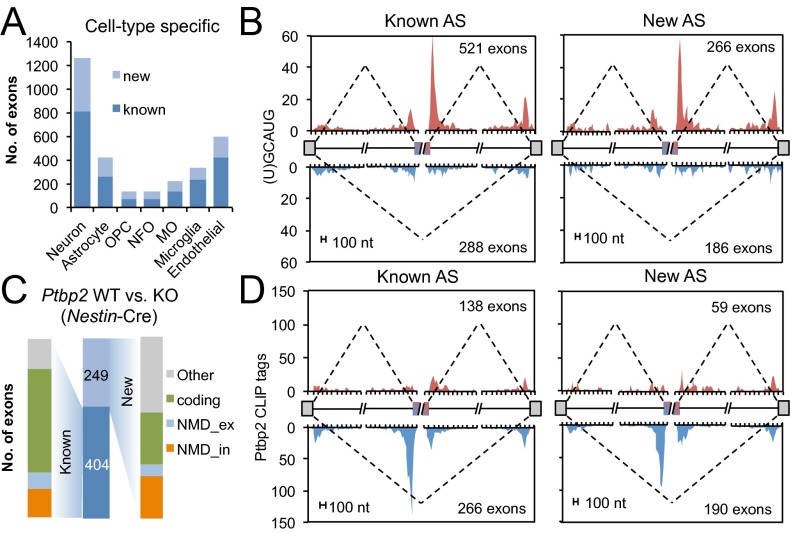
Fig. 3.
Known and new cassette exons under tissue- or cell type-specific regulation. In each panel, only nonredundant cassette exons were included for analysis. (A) The number of known and new cassette exons with cell type-specific AS. MO, myelinating oligodendrocyte; NFO, newly formed oligodendrocyte; OPC, oligodendrocyte precursor cells. (B) An RNA map correlating neuron-specific exon inclusion (red) or exclusion (blue) with the position of (U)GCAUG elements recognized by Rbfox proteins. (C) The number of known and new cassette exons with altered splicing upon CNS-specific depletion of Ptbp2 (driven by Nestin-Cre, middle bar). For each group, exons are further classified with respect to their alternative coding capacity. (D) An RNA map correlating Ptbp2-dependent exon inclusion (red) or exclusion (blue) with the position of Ptbp2 binding as measured by Ptbp2 CLIP tags.