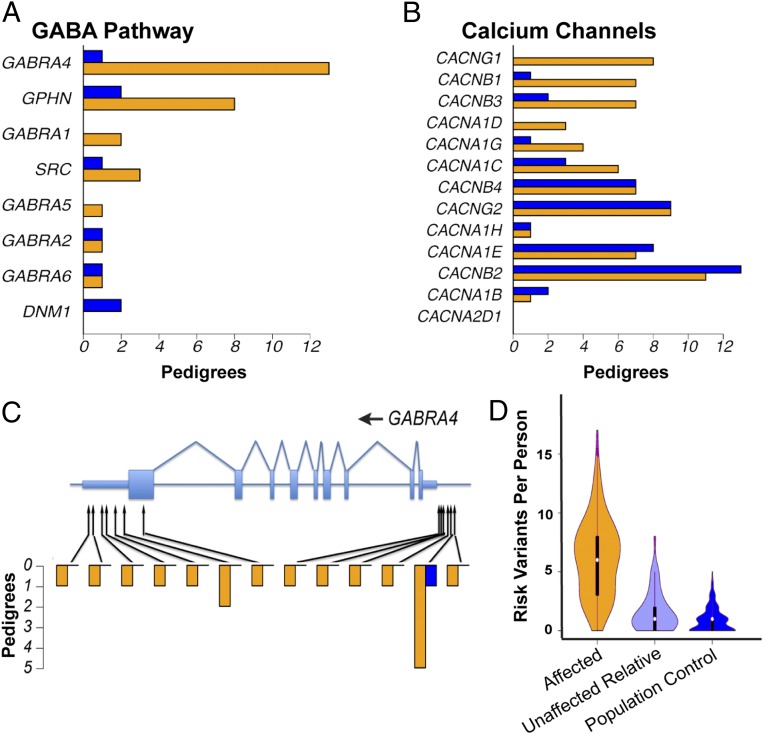
Fig. 2.
Pathways enriched for rare variants in BD pedigrees. (A) Number of BD (orange) and control (blue) pedigrees in which a fully or nearly fully segregating coding or regulatory variant was found in a gene assigned to the GABA pathway by BioCarta. The 34 control pedigrees used in this analysis were of European ancestry and were matched for similar size and structure to the BD pedigrees. (B) Number of BD (orange) and control (blue) pedigrees in which a fully or nearly fully segregating coding or regulatory variant was found in a gene assigned to the Gene Ontology term “Voltage-Gated Calcium Channel Complex.” (C) Fully and nearly fully segregating coding and regulatory SNVs and indels in the GABA receptor gene GABRA4. Protein-coding regions and UTRs are shown, respectively, by wide and narrow light blue rectangles. Heights of orange and blue bars indicate the number of BD and control pedigrees in which each variant was fully or nearly fully segregating. (D) Number of risk variants identified in each BD case (orange), unaffected individual in a BD pedigree (light blue), or population control (dark blue). For this analysis, we defined as risk variants all SNVs with P < 0.001 and odds ratios > 1 by mixed model analysis (14 SNVs), as well as SNVs with mixed-model P < 0.05, odds ratios > 1, and an annotation to one of the following pathways with an increased burden of rare variants in BD pedigrees (SI Appendix, Table S4): BioCarta GABA pathway (9 SNVs), voltage-gated calcium channel complex (38 SNVs), CaM kinases (10 SNVs), GTPases (88 SNVs), and glycolysis/tricarboxylic acid cycle (13 SNVs). The widths of polygons are proportional to the number of individuals with each variant count.