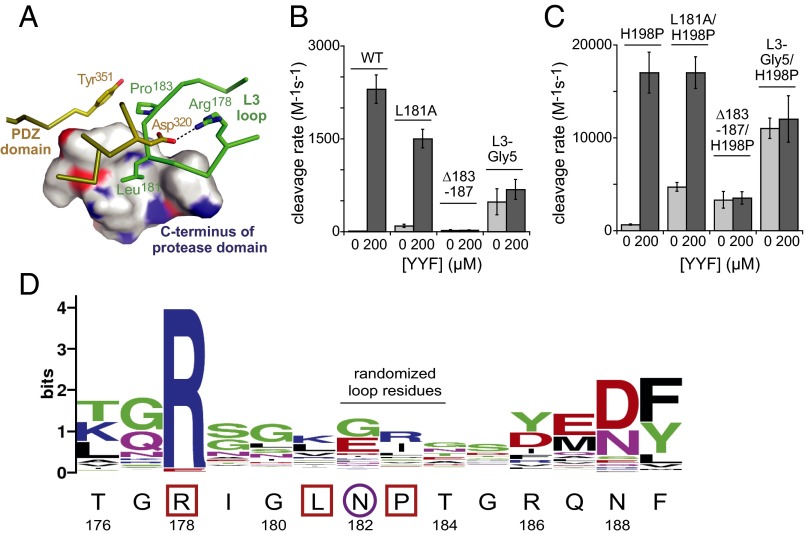
Fig. 5.
The L3 loop. (A) Autoinhibitory interactions involving L3-loop residues (green) stabilize the inactive conformation of DegS (PDB ID: 4RR1). The side chain of Leu181 packs into a hydrophobic pocket formed by Leu240, Ile244, Lys247, and Val254 from the C-terminal part of the protease domain, and a hydrogen bond between the Arg256 side chain and the carbonyl oxygen of Leu181 helps stabilize these interactions. In addition, Arg178 makes a salt-bridge with Asp320 in the PDZ domain, and Pro183 packs against Tyr351 in the PDZ domain. (B) Basal and YYF-stimulated cleavage activities of wild-type DegS and variants with L3-loop mutations. (C) Basal and YYF-stimulated cleavage activities of H198P DegS and variants with L3-loop mutations. (D) Most residues in the L3 loop are poorly conserved in an alignment of prokaryotic and eukaryotic proteases homologous to DegS. The wild-type DegS sequence is shown below the WebLogo figure (19). L3-loop residues that mediate DegS autoinhibition are boxed; residue 182, which is critical for linking OMP binding to allosteric activation, is circled. Asn182 is flanked by residues that make autoinhibitory contacts, restricting its ability to avoid a clash upon OMP binding to inactive DegS.