Fig. 5.
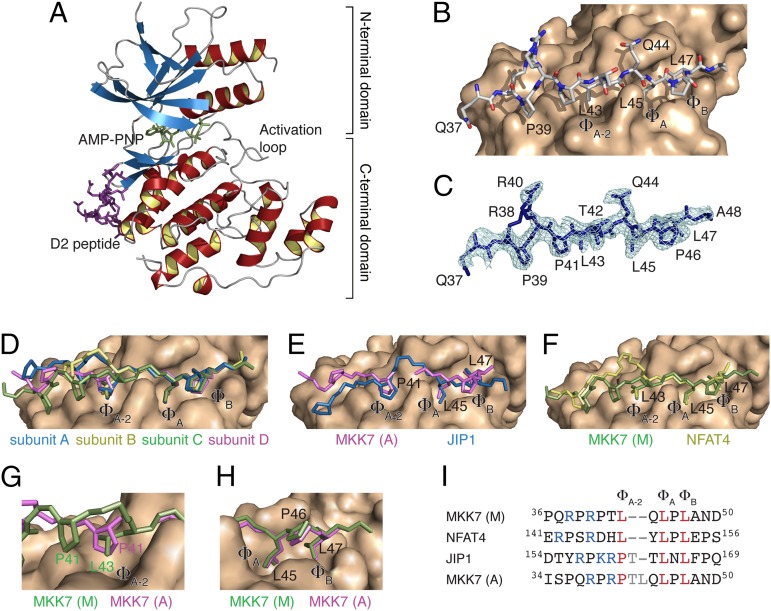
Crystal structure of JNK1 in complex with the D2 docking site peptide. (A) Crystal structure of JNK1 in complex with the ATP analog AMP-PNP (green) and the D2 peptide (magenta). (B) Structural basis of the interaction between the D2 peptide (sticks) and JNK1 (surface) as observed in chain C (main binding mode). (C) Simulated annealed omit electron density map contoured at 1σ of the D2 peptide of chain C. (D) Superposition of the four MKK7 docking site peptides on the surface of JNK1. The MKK7 peptides are shown as sticks with side chains displayed for residues P41, L43, L45, P46, and L47. (E) Superposition of the MKK7 (magenta, alternative binding mode) and JIP1 (blue) docking site peptides on the surface of JNK1. (F) Superposition of the MKK7 (green, main binding mode) and NFAT4 (yellow) docking site peptides on the surface of JNK1. (G) Zoom of the ΦA-2 hydrophobic pocket. (H) Zoom of the ΦA and ΦB hydrophobic pockets. (I) Sequence alignment of the docking site motif of MKK7 with that of NFAT4 and JIP1. Two alignments are shown for MKK7 corresponding to the main (M) and alternative (A) binding mode observed in the crystal structure.