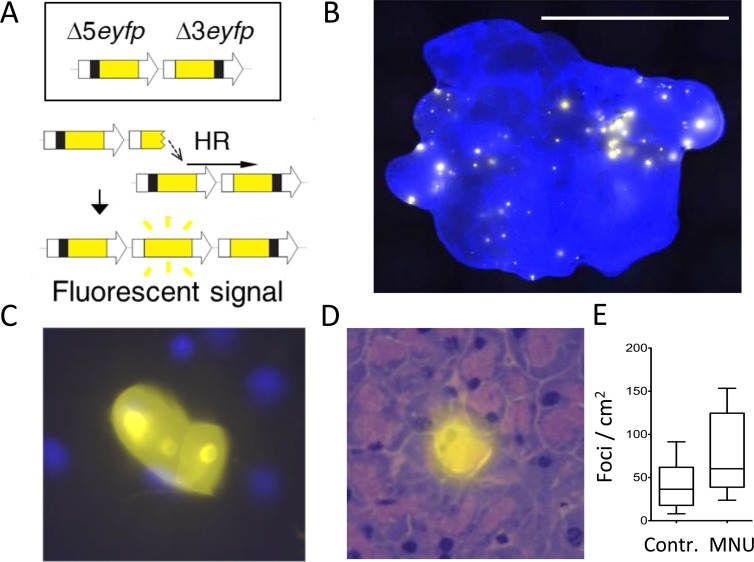
Figure 1. The FYDR mouse detects HR-derived sequence rearrangements in situ in intact tissue.
(A) Schematic of the reconstitution of full-length EYFP coding sequence from two truncated copies through replication fork restart by HR. Note that the appearance of fluorescent signal indicates the gain of one repeat unit (a duplication). Arrows represent expression constructs. EYFP coding sequences are in yellow, promoter and polyadenylation signal sequences are in white, and deleted sequences are in black. Drawing is not to scale. (B) Representative image of a FYDR pancreas showing fluorescent foci detectable in situ in intact tissue. Freshly harvested, unfixed whole pancreas was counterstained with Hoechst, compressed to 0.5 mm and imaged under an epifluorescent microscope. Fluorescence is pseudocolored. Original magnification, ×1. Scale bar = 1 cm. (C) Cluster of recombinant cells at ×60 original magnification. Fluorescence is pseudocolored. (D) A recombinant pancreatic acinar cell identified by the overlay of EYFP fluorescence and H&E staining. Fluorescence is pseudocolored. Original magnification, ×40. (E) The model alkylating agent MNU induces HR in the pancreas. Mice received 25 mg/kg MNU i.p., and HR was evaluated 3 to 5 weeks after treatment. Frequencies of recombinant foci per cm2 tissue area are significantly greater in MNU-treated mice (n = 15) than in control mice (n = 16). Boxes show 25th and 75th percentiles, medians are indicated by horizontal lines. * P < 0.05 (Mann–Whitney U-test).