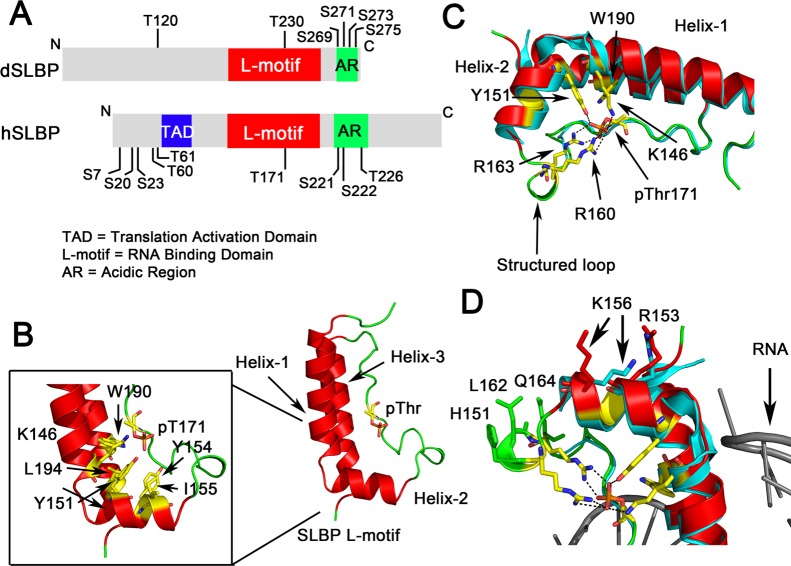
Figure 3.
X-ray crystal structure of phosphorylated human SLBP. (A) Schematic showing the domain organization of human SLBP (hSLBP) and Drosophila SLBP (dSLBP). The RNA binding domain is designated the “L-motif” and is followed by an acidic region, rich in Asp and Glu residues, that is also phosphorylated. The N-terminal domain is involved in translation activation (TAD). Phosphorylation sites that have been mapped in vivo(76,80,124) are indicated. (B) The T171 phosphorylated SLBP L-motif is shown with a characteristic L-shape as seen in the crystal structure of the hSLBP/histone mRNA/3′hExo ternary complex (PDB code 4QOZ). The fold consists of three α-helices connected by a 20-residue flexible loop that has the site of phosphorylation (shown in stick). Hydrophobic residues at the junction of the helices are shown in yellow (inset). (C) Hydrogen bonding interactions mediated by the phosphothreonine with R163, R169, K146, Y151, and W190 (via a water molecule) are shown. The structured loop that is disordered in the unphosphorylated SLBP structure is fully ordered in phosphorylated SLBP. The unphosphorylated structure is shown in blue and the phosphorylated structure in red ribbon. (D) Residues in helix-2 and the structured loop that undergo a conformational change upon SLBP phosphorylation and have been implicated in RNA processing are highlighted.