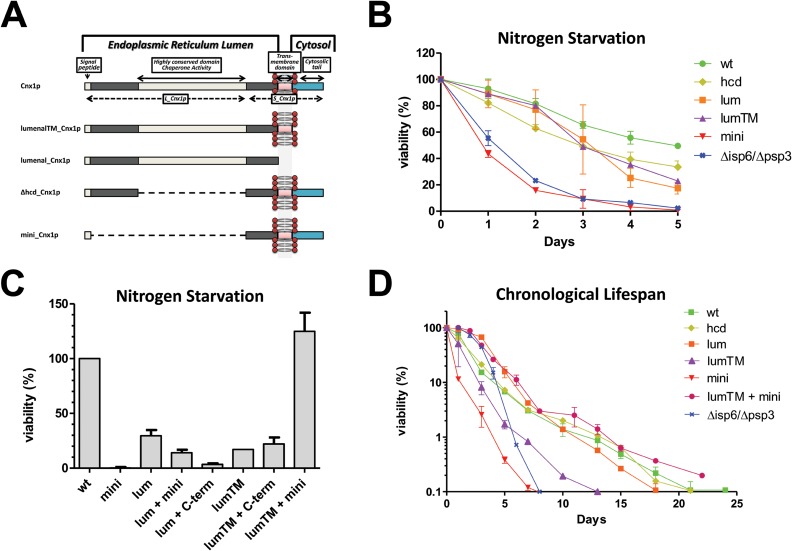
Fig 5. Different regions of calnexin are required for survival under nitrogen starvation and in stationary phase.
(A) Schematic representation of wild-type calnexin (Cnx1p) and truncated mutants. The Large (L_Cnx1p) and small domain (S_Cnx1p) obtained after the processing of calnexin are depicted in the full length version. (B) Viability under nitrogen starvation. Strains mini_cnx1 (SP18344), lumenal_cnx1 (18346), lumenalTM_cnx1 (SP18348), Δhcd_cnx1 (SP18350), isp6Δpsp3Δ (SP18340) and WT control cells (SP18342) were grown in EMM until early exponential phase (OD600<0.5), washed twice with water and resuspended in EMM-N. Culture samples were plated every 24h onto EMM, and incubated for 5 days at 30°C. CFU/ml was determined and 100% viability was set as the maximum CFU/ml reached for each strain before decline (usually 24h after the shift). (C) Co-expression of lumenalTM_Cnx1p rescues the rapid loss of viability of mini_cnx1 under nitrogen depletion. The viability of the strains, presented as relative to WT defined as 100%, mini_cnx1 (SP18344), lumenal_cnx1 (SP18346), lumenal_cnx1 + mini_cnx1 (SP18282), lumenal_cnx1 + C-term_cnx1-Venus (SP18300), lumenalTM_cnx1 (SP18348), lumenalTM_cnx1 + C-term_cnx1-Venus (SP18303), lumenalTM_cnx1 + mini_cnx1(SP18285) and control cells (SP18342) was analyzed as described in (B). (D) Analysis of chronological lifespan (CLS) in calnexin mutants. Strains mini_cnx1 (SP18344), lumenal_cnx1 (SP18346), lumenalTM_cnx1 (SP18348), Δhcd_cnx1 (SP18350), lumenalTM_cnx1 + mini_cnx1 (SP18285), isp6Δpsp3Δ (SP18340) and WT control cells (SP18342) were cultured in EMM supplemented with all the amino acids, except those required for selection (EMMC). Culture samples from different days (within a period of 25 days) were plated onto EMM and incubated at 30°C for 5 days. CFU/ml was determined and 100% was set as the maximum CFU/ml reached for each strain. Y-axis is shown in logarithmic scale.