Abstract
The question as to why parasites remain generalist or become specialist is a key unresolved question in evolutionary biology. Ampelomyces spp., intracellular mycoparasites of powdery mildew fungi, which are themselves plant pathogens, are a useful model for studies of this issue. Ampelomyces is used for the biological control of mildew. Differences in mycohost phenology promote temporal isolation between sympatric Ampelomyces mycoparasites. Apple powdery mildew (APM) causes spring epidemics, whereas other powdery mildew species on plants other than apple cause epidemics later in the season. This has resulted in genetic differentiation between APM and non-APM strains. It is unclear whether there is genetic differentiation between non-APM Ampelomyces lineages due to their specialization on different mycohosts. We used microsatellites to address this question and found no significant differentiation between non-APM Ampelomyces strains from different mycohosts or host plants, but strong differentiation between APM and non-APM strains. A geographical structure was revealed in both groups, with differences between European countries, demonstrating restricted dispersal at the continent scale and a high resolution for our markers. We found footprints of recombination in both groups, possibly more frequent in the APM cluster. Overall, Ampelomyces thus appears to be one of the rare genuine generalist pathogenic fungi able to parasitize multiple hosts in natural populations. It is therefore an excellent model for studying the evolution of pathogens towards a generalist rather than host-specific strategy, particularly in light of the tritrophic interaction between Ampelomyces mycoparasites, their powdery mildew fungal hosts and the mildew host plants.
Introduction
The question of why parasites remain generalist or become specialist remains a key unresolved issue in evolutionary biology. Generalist parasites have access to a larger number of individual hosts, but there may be trade-offs impeding optimal adaptation to several host species at the same time [1–3]. These trade-offs may take the form of constraints or a lower speed of coevolution when tracking a particular host species [4]. Specialist species may therefore be at an advantage, as they should exploit their hosts more efficiently, but this is likely to be the case only if the principal host is sufficiently abundant [5]. For specialization to occur, there must also be a mechanism preventing gene flow between sympatric parasite species adapting to different hosts, to avoid the break-up of adaptive combinations of alleles [6–8]. If this condition is not fulfilled, specialization cannot evolve and parasites remain generalists on multiple sympatric hosts. The mechanisms underlying reproductive isolation in parasites include mate choice and intersterility [9], as in free-living organisms, but also temporal isolation due to differences in host phenology [10, 11], mating within hosts after growth [12–15] and selfing [8, 16].
It is therefore not straightforward to predict whether and when a parasite will remain generalist or evolve towards specialization. Many case studies are required before we can draw general inferences. True generalist parasites appear to be rare; morphological species long considered generalist are often found to be cryptic specialist species in studies based on molecular markers [8, 17, 18]. However, genuine generalist parasites do exist, and this group includes the fungi Botrytis cinerea, Sclerotinia sclerotiorum, and Batrachochytrium dendrobatidis [19–22].
Geography is another driver of genetic differentiation in parasites. Spatial genetic structure has been detected in a number of parasites, even in fungi with wind-borne spores [15, 23–29]. Spatial genetic structure can be used to infer dispersal distances, which is useful for the monitoring and prediction of epidemics caused by pathogenic fungi [23, 24, 30–32]. In this respect, however, our current knowledge on fungal pathogens is relatively limited, especially in the case of mycoparasites, i.e., fungi parasitizing other fungi, with only a few studies available, on tremellalean mycoparasites [33] and some lichenicolous fungi [34, 35].
Ampelomyces is an interesting biological model in this concept. These fungi are intracellular mycoparasites of powdery mildew fungi, which are themselves plant pathogens [36, 37]. Differences in host phenology promote temporal isolation between sympatric mycoparasites infecting apple powdery mildew (Podosphaera leucotricha) and those infecting other powdery mildew species on host plants other than apple [11]. Indeed, apple powdery mildew (APM) epidemics occur in spring, whereas the fungal hosts of the other Ampelomyces species cause epidemics mostly in the summer and fall. Consequently, Ampelomyces populations infecting APM display strong differentiation from mycoparasites infecting other mildews, despite their ability to infect these other mildews in laboratory and field experiments [11]. However, it was not possible to investigate genetic differentiation between the Ampelomyces parasites of different mildews species causing epidemics in summer and fall in these previous studies. Indeed, the microsatellite markers developed for the mycoparasites of apple powdery mildew gave no amplification in Ampelomyces strains isolated later in the season, because the level of genetic differentiation was too high. Gene sequences revealed no clear separation according to host species in non-APM Ampelomyces [38–41] but differentiation may have occurred too recently for detection by this approach.
No strong geographic structure was found in our previous study focusing on European Ampelomyces strains from apple powdery mildew [11]. Microsatellite markers revealed long-distance spread across Europe, consistent with the wind dispersal of these mycoparasites within airborne parasitized powdery mildew spores (Szentivanyi & Kiss 2003; Kiss 2008). However, further knowledge of the geographical structure and dispersal ability of Ampelomyces populations is required, especially because Ampelomyces strains are sold as biological control agents [42].
Another interesting question in Ampelomyces is the extent of recombination. Sexual structures have never been observed in the field, but some footprints of recombination have been detected in Ampelomyces from apple powdery mildew [11]. Recombination traces have proved to be common in fungal species previously thought to be asexual [43–51], reflecting rare sexual events or the occurrence of sex in places or at times not accessible to observation. We were unable to investigate the occurrence of sex in Ampelomyces from hosts other than apple powdery mildew, because of the lack of polymorphic markers [11].
We report here the development of new microsatellite markers for mycoparasites isolated from powdery mildew fungi in the summer and fall, and their use to genotype a large collection of mycoparasites from a number of powdery mildew species and host plants, worldwide but with a particular effort in Europe. We addressed the following questions: 1) Is there any genetic differentiation between Ampelomyces strains isolated from different host plants affected by mildew epidemics in the summer and fall that might indicate specialization? 2) Is there any geographic structure within Ampelomyces? 3) Are there footprints of recombination in Ampelomyces, within and between powdery mildew hosts?
Materials and Methods
Fungal materials
We studied 469 Ampelomyces strains maintained in culture and 168 dried powdery mildew-infected leaf samples containing Ampelomyces pycnidia (S1 Table). Leaf samples were obtained as described by Kiss et al. (2011) and each was treated as a single Ampelomyces strain (haplotype) on the basis of microsatellite genotyping results [52]. We thus studied 637 Ampelomyces strains in total. Twelve of these strains were newly obtained from Blumeria graminis, the only powdery mildew species known to infect various monocot species. These new strains were isolated as described previously [40]. The rest of the strains came from previous works [11, 38, 40, 41, 53–55] (S1 Table). Most of these strains, 157 in total, came from Arthrocladiella mougeotii, a powdery mildew species infecting a solanaceous weed, Lycium halimifolium. A group of 17 strains were isolated from Erysiphe necator (formerly Uncinula necator) infecting grapevine. The remaining strains came from powdery mildew species infecting various host plant species, none of which was widely sampled. Overall, our dataset of 637 Ampelomyces strains contained 394 strains infecting apple powdery mildew (APM) and 243 non-APM strains. We chose B. graminis, A. mougeotii and E. necator to be the most intensively sampled non-APM mycohost species because these three powdery mildews have been shown to be representative of the diversity of non-APM species [56].
DNA extraction
Total genomic DNA was extracted from about 10 to 15 mg of freeze-dried mycelium for each Ampelomyces strain and from about 5 to 10 mg of dried leaf sample for samples containing Ampelomyces pycnidia. We used the DNeasy Plant Mini Kit (Qiagen) for DNA extraction.
Development of microsatellite markers
Microsatellite markers were identified as described previously [52], using 11 strains (highlighted in S1 Table) isolated in the summer or fall, from different mycohosts in Europe, Israel and China. They belonged to different ITS and actin gene clades [38]. The genotypes of all the strains tested in this work are given in S2 and S3 Tables.
Population genetic analyses
Microsatellite genotyping. Amplifications were carried out with forward primers labeled with fluorescent FAM or HEX dyes (Sigma Aldrich). Each PCR was performed in a final volume of 10 μl containing 5 μl Multiplex PCR Master Mix (Qiagen) and 0.2 μl of each primer at a concentration of 10 μM and 2 μl of genomic DNA. The primer sequences are shown in Table 1. Three primer combinations were used: set 1 (LK3c and LK7d), set 2 (LK3a, LK7c and LK10c) and set 3 (LK3b, LK7f and LK10d). Assays were carried out in the following conditions: initial denaturation for 5 minutes at 95°C, followed by 28 cycles of denaturation for 45 s at 95°C, annealing for 50 s at 54°C, extension for 1 minute at 72°C, and a final extension for 5 minutes at 72°C. PCR products were heated at 94°C for 5 minutes, chilled on ice and separated on an ABI Prism 3130XL Genetic Analyzer (Applied Biosystems). Each gel contained GenScan-500LIZ Size Standards (35 to 500 bp, Applied Biosystems), for the analysis of microsatellite data with GeneMapper Software (Applied Biosystems).
Table 1. Characteristics of the eight polymorphic microsatellite loci identified in Ampelomyces strains isolated from many different mycohosts in autumn.
| Locus | Motif | Primer sequence (5'-> 3') | Allele size range | No. ofalleles |
|---|---|---|---|---|
| LK3a | [CAGCAGCA-n15]8 | CAAGATCTGCCGCCAACC | 203–302 | 25 |
| GGTGTTGTTGTGCATGTTGTC | ||||
| LK3b | [cgg]5 | CGGCACGAAATCTACCTGTC | 126–144 | 5 |
| CGCAGAGGTGGATGTAGGTT | ||||
| LK7c | [ggt/gga/ggc]3 | GATAGGACGCGGTTATGGAA | 176–221 | 9 |
| GATGTGGCTTCCTGGTTCG | ||||
| LK10c | [gct]6[cgg]3 | CATCGTGATGTTCTGGGTGA | 149–197 | 10 |
| AGACCACAATCTCCGACCAG | ||||
| LK10d | [ggt]5[ggcggt]3 | CAGAAGTGGATTGCGGAGAG | 179–257 | 21 |
| CAAGGCCACATCCAAGTTCT | ||||
| LK3c | [aag]13 | GACAAGAAACCTTGGGTTGG | 101–161 | 20 |
| GGACGACGATTTGCAGACTA | ||||
| LK7d | [caccgc]4 | GCTTCGGGTTTGTCTCAGTC | 146–188 | 7 |
| CGAAAGGGTTGATGAGGTTT | ||||
| LK7f | [gt]12 | AAAAGTCAAGGGACCACACG | 181–253 | 23 |
| CATAAGCGATGGGAGTTGGT |
The first five loci listed below were also used to genotype Ampelomyces strains isolated from apple powdery mildew (APM) in spring.
Genetic analyses: SplitsTree, correspondence analyses, linkage disequilibrium, and structure. SplitsTree 4 [57] (http://www.splitstree.org/) was used to visualize differentiation and the occurrence of recombination, (i) for the total dataset, with the five microsatellite markers yielding amplification products for all samples, and (ii) on the 209 non-APM strains that could be genotyped with all eight markers. Factorial correspondence analyses (FCA) were performed with GENETIX v4.05 [58]. FST and linkage disequilibrium between the five markers and population differentiation were assessed online with Genepop [59, 60], http://genepop.curtin.edu.au/. We used the Genclone software [61] to determine the number of different multilocus genotypes (MLGs) in the datasets and for the estimation, by permutation, of the expected number of MLGs for different numbers of markers.
We used the individual-based Bayesian clustering methods implemented in STRUCTURE 2.3.3 [62] to infer population structure. STRUCTURE makes use of Markov Chain-Monte Carlo (MCMC) simulations to infer the proportion of ancestry of genotypes from K distinct clusters. The underlying algorithms attempt to minimize deviations from Hardy–Weinberg and linkage disequilibria. Ten independent analyses were carried out for each number of clusters, from K = 2 to K = 10, with admixture models and 500,000 MCMC iterations, after a burn-in of 500,000 steps. Outputs were processed with CLUMPP v1.1.2 [63], to identify clustering solutions in replicated runs for each value of K. Population structure was then displayed graphically with DISTRUCT v1.1 [64]. We used the Evanno method, via the STRUCTURE Harvester website (http://taylor0.biology.ucla.edu/structureHarvester/) [65], to identify the K value corresponding to the strongest structure.
Results
Development and characteristics of new microsatellite markers
We identified and developed eight new microsatellite markers (Table 1) using 11 Ampelomyces strains isolated from different mycohosts during the fall (S1 Table). Five of these eight new markers, LK7c, LK3a, LK10c, LK3b and LK10d, amplified the target loci in all the APM and non-APM strains studied (S2 Table), regardless of the season in which they were isolated. The number of alleles was found smaller in APM strains than in non-APM strains (Table 2). The remaining three markers could be amplified from only 209 strains, all of which were non-APM strains belonging to different lineages according to their ITS and actin gene sequences [38–40]. Most of these phylogenetically diverse strains were isolated in summer or fall, in different parts of the world (China, France, Germany, Hungary, Israel, Italy, Korea, South Africa, the United Kingdom and the USA); the season in which isolation occurred was not known for a few strains (S1 Table).
Table 2. Number of alleles detected at the five microsatellite markers in 394 Ampelomyces strains isolated in spring from apple powdery mildew (APM strains), and in 243 non-APM strains isolated later in the season from many other powdery mildew species infecting various host plants.
| Strain type | Locus | ||||
|---|---|---|---|---|---|
| LK7c | LK3a | LK10c | LK3b | LK10d | |
| APM strains | 1 | 5 | 4 | 2 | 3 |
| non-APM strains | 8 | 20 | 7 | 3 | 18 |
The microsatellite profiles of the strains are shown in S2 Table.
Genetic diversity and population structure in Ampelomyces strains
We first investigated the population structure of Ampelomyces strains with STRUCTURE [66]. SplitsTree analyses were also carried out, separately for the genotypes of all the strains based on five microsatellite markers (Fig. 1), and for the 209 non-APM strains genotyped with the eight markers (Fig. 2). STRUCTURE yielded well defined clusters at K values of up to 6 (Fig. 3 and S1 Fig.), indicating the existence of six genetically differentiated clusters. For values of K of 7 and above, each new cluster included only admixed genotypes, indicating a lack of further structure. The deltaK value [67] confirmed that the split into six populations corresponded to a peak in the strength of the structure in the dataset, the strongest peak being at K = 4 (S2 Fig.). At K = 2, apple powdery mildew (APM) strains were separated from all other (non-APM) strains. This differentiation between APM and non-APM strains also appeared clearly on the SplitsTree (Fig. 1). At K = 6, non-APM strains were split into two clusters (Fig. 3, populations 1 and 2), whereas strains isolated from apple powdery mildew were split into four clusters (Fig. 3, populations 3 to 6). The clusters of non-APM strains did not correspond to either different host mildews or different host plants, but some geographical clustering was revealed: one of the clusters encompassed most of the Hungarian strains (128 of 130), together with two Italian strains, whereas the second cluster contained strains of more diverse origins, from Hungary, France, the United Kingdom, China, the USA, and Israel. Some geographical structure was also observed within APM strains: populations 5 and 6 mostly included strains from France, together with 13 of the 104 strains from Hungary (in population 5), and 3 of the 60 strains from the UK (in the population 6), whereas populations 3 and 4 included strains from different parts of Europe (Hungary, France, Germany, and the UK). On the other hand, the non-European strains appeared scattered in the SplitsTree (arrows on Fig. 2).
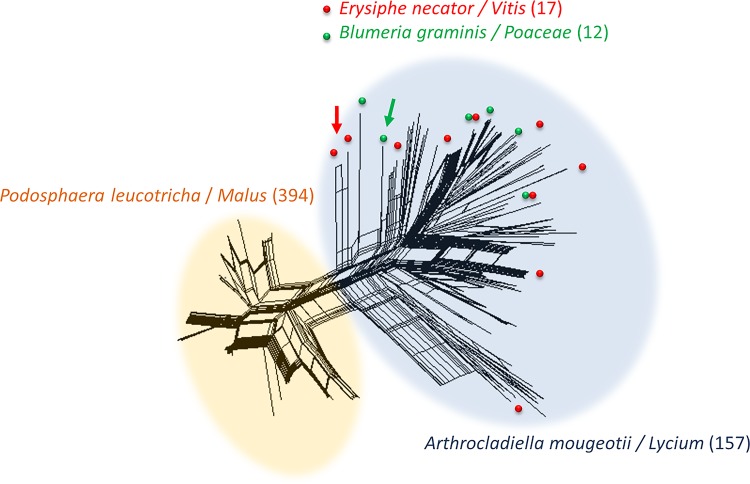
Fig 1. SplitsTree analysis of all 637 Ampelomyces strains, based on five microsatellite markers.
Widely sampled fungal hosts and the plant hosts from which they were collected are indicated by colors and the number of strains is indicated in brackets. Two main clusters were identified, one consisting of the 394 strains from apple powdery mildew (APM, Podosphaera leucotricha, light orange cloud), isolated in spring, and the other comprising all the non-APM strains, i.e. the 157 strains isolated from Arthrocladiella mougeotii infecting Lycium halimifolium plants in Hungary (light blue cloud) and the strains isolated from other powdery mildew hosts in Europe and elsewhere, later in the season. In this second cluster, positions are shown by arrows for the 17 strains isolated from grapevine powdery mildew (Erysiphe necator, in red) and the 12 strains isolated from Blumeria graminis infecting wild grass species (in green). Reticulation indicates the occurrence of recombination.
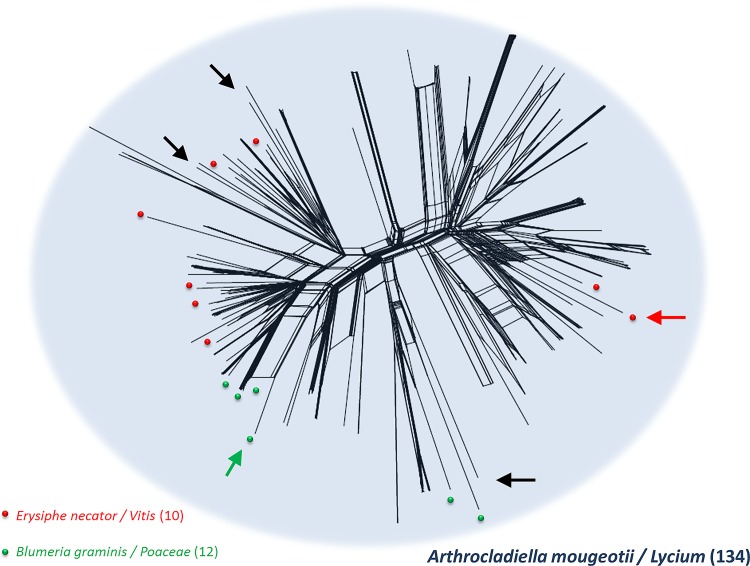
Fig 2. SplitsTree analysis of the 209 non-APM Ampelomyces strains in which the eight microsatellite markers could be amplified.
Widely sampled fungal hosts and the plant hosts from which they were collected are indicated by colors and the number of strains is shown in brackets: 134 strains came from Arthrocladiella mougeotii infecting Lycium halimifolium plants (blue cloud), 12 strains came from Blumeria graminis infecting grasses (green points), 10 strains from grapevine powdery mildew (Erysiphe necator) (red points), and 53 other strains were isolated from several other powdery mildew species infecting various host plants, isolated in summer and fall. The position of an Ampelomyces strain isolated from E. necator in the USA and that of another strain isolated from B. graminis in Korea are indicated by red and green arrows, respectively. Black arrows indicate the positions of the other three non-European strains. Reticulation indicates the occurrence of recombination.
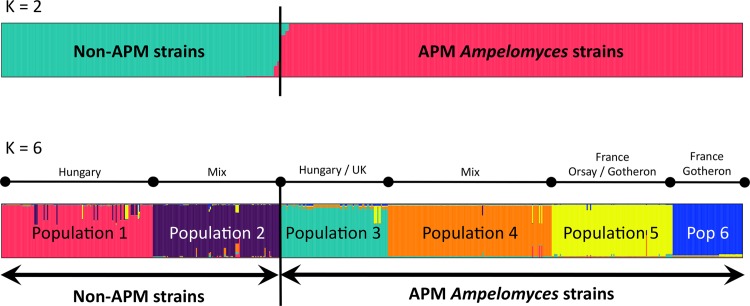
Fig 3. Population structure in Ampelomyces.
Structure was inferred by STRUCTURE for K = 2 and K = 6 (see S1 Fig. for the bar plots corresponding to other K values). STRUCTURE yielded well defined clusters up to K = 6, indicating the existence of six genetically differentiated clusters. K = 2 separates APM and non-APM strains. The geographical origin of strains is indicated on the K = 6 barplots.
The existence of six genetically divergent populations was confirmed by FST analyses. The fixation index was 0.25 between the non-APM populations (populations 1 and 2), indicating strong genetic differentiation. Pairwise fixation indices ranged between 0.22 and 0.42 between the other four populations, indicating that there was also strong genetic differentiation between APM populations. The level of genetic differentiation was strongest between APM and non-APM clusters, with an FST value of 0.49 between the two clusters obtained at K = 2.
A factorial correspondence analysis (FCA) yielded further support for the genetic patterns revealed above (S3 Fig.), with 7.82%, and 4.38% of the variance explained by axes 1 and 4, respectively). APM strains and non-APM strains appeared to be strongly separated along axis 1. The other populations were also segregated, but less strongly.
Recombination footprints
In the dataset based on five markers and 637 strains, only 93 MLGs were detected, with a mean of 6.7 strains per MLG. These identical MLGs probably did not correspond entirely to clones, resulting instead from the insufficiently high discriminatory power of the five markers, as a permutation estimating the number of MLGs as a function of the number of markers used did not reach a plateau (S4 Fig.); actually, most of these identical MLGs could be differentiated using eight markers. In the dataset based on eight markers and 209 strains, 91 MLGs were detected, with a mean of only 2.3 strains per MLG, and a single abundant MLG for 17 strains in the APM cluster. In this analysis, the number of MLGs as a function of the number of markers used did begin to reach a plateau (S4 Fig.), although the shape of the curve indicated that discrimination could probably be increased further by the use of a larger number of markers. Thus, the identical MLGs obtained probably do not strictly correspond to clones and did not, therefore, bias the structure analyses.
We assessed the occurrence of sex, by looking for footprints of recombination in the split decomposition analysis, in which recombination events were identified as reticulations (Figs. 1 and 2). The SplitsTree analyses yielded networks with long branches, but also with some reticulations, mostly within each of the APM and non-APM clusters, with a smaller number between APM and non-APM strains. This pattern is indicative of pervasive clonality with occasional recombination events, these events being less frequent between than within the APM and non-APM clusters, confirming that differences in host plant phenology impede gene flow between these two groups. The branches appeared to be shorter and the reticulations more frequent within the APM cluster than within the non-APM cluster (Fig. 1), confirming the lower level of diversity and suggesting less ancient clonality and/or more frequent recombination events. The SplitsTree analyses have also shown that there was no detectable genetic differentiation within non-AMP strains as a function of the powdery mildew host, as strains from different powdery mildew species were scattered (Figs. 1 and 2). Non-APM strains belonging to different phylogenetic lineages based on their ITS and actin gene sequences [38–40] did not form distinct groups in any of the SplitsTree analyses (Figs. 1 and 2).
Linkage disequilibrium analyses were consistent with the SplitsTree and STRUCTURE analyses, indicating frequent clonality with occasional recombination events, these events being more frequent in the APM cluster. Linkage disequilibrium was, indeed, significant within both AMP and non-APM clusters, at least partly reflecting a Wahlund effect due to the genetic structure. When the populations were considered separately, we also detected significant linkage disequilibrium within both the non-APM populations (populations 1 and 2), whereas no significant linkage disequilibrium was detected for the three APM populations for which linkage disequilibrium could be calculated.
Discussion
In this study, we developed eight new polymorphic microsatellite markers for investigating the population structure and diversity of Ampelomyces strains isolated from many powdery mildew species infecting very different host plant species and representative of the genetic diversity in Erysiphales. Five out of eight of our newly developed microsatellite markers could be amplified in both AMP and non-AMP strains, which was not the case for previous microsatellite markers [11]. The newly developed markers will be useful for detecting and monitoring Ampelomyces strains released as commercial biological control agents in vineyards [42] and in agricultural fields, because they were amplified reliably from environmental samples as well as from strains maintained in culture.
Various analyses, using a comprehensive dataset with the genotypes of APM and non-APM strains, confirmed that there was a high level of genetic differentiation between these two groups, with no evidence of gene flow. This is consistent with the findings of our previous study, indicating that APM strains form a distinct cryptic species, genetically isolated from non-APM strains by temporal isolation due to differences in the phenology of their powdery mildew hosts [11]. Indeed, apple powdery mildew overwinters in apple buds [41] and begins to sporulate soon after bud burst, becoming widespread on its host plants in spring and persisting without much further spread, mostly on green shoots, during the summer and fall. By contrast, the other powdery mildew species, the mycohosts of the non-APM strains, begin to sporulate and spread on their respective host plants later in the season. They also continue to display active sporulation and spread on host plants until late fall [11]. Phylogenetic analyses have previously suggested that the non-APM strains could be classified into distinct molecular taxonomic units (MOTUs), which were not specific to particular mycohost or plant host species [38–40, 53]. Neither SplitsTree nor STRUCTURE analyses however confirmed here the distinction of the non-APM strains according to the MOTUs defined in these previous works [38–40, 53].
STRUCTURE analyses further revealed that the Ampelomyces strains included in this work could be split into six distinct populations (Fig. 3). The non-APM strains formed two genetically differentiated populations that could be accounted for by geography, as one of the clusters contained mostly Hungarian strains. This differentiation of Hungarian strains probably reflects their abundance in the dataset, and further geographical clustering may be revealed within the other clusters after additional sampling. The APM strains formed four different populations that also displayed some geographical clustering. Most of the strains, both APM and non-APM, came from Hungary and two regions of France, again potentially explaining why STRUCTURE differentiated these three regions. This study provides the first evidence of geographical structure in Ampelomyces, confirming that, even in wind-dispersed fungi “not everything is everywhere”, and that gene flow is restricted by distance, as shown in many other fungi [15, 23–29]. The ability of our markers to reveal geographical structure in European populations highlights their discriminatory power and indicates that the lack of structure according to host is real and not due to low power.
The analyses of non-APM Ampelomyces populations detected no structure according to powdery mildew mycohost and/or host plant species, even in the most widely sampled areas. Moreover, strains isolated from different powdery mildew species, infecting different host plants, sometimes had identical, or very similar, microsatellite profiles. These results suggest that there are no barriers restricting gene flow among Ampelomyces strains affecting different powdery mildew species, on different host plants, in the same environment. These findings suggest that this mycoparasite is a genuine generalist. This may appear contrary to the results of laboratory experiments showing that Ampelomyces strains from grapevine powdery mildew parasitize their original mycohost species more strongly than two other test powdery mildew species [55], or showing differences in the virulence of Ampelomyces strains against three powdery mildew species [68]. However, there may be some polymorphism in the ability of different strains to infect various host species or aggressiveness towards different host species within a given mycoparasite species. By contrast, other cross-inoculation experiments have repeatedly shown that a number of Ampelomyces strains isolated from different powdery mildew species were all able to parasitize test powdery mildew species in the laboratory [40, 41, 69] with similar intensities of mycoparasitism for strains isolated from conspecific and other powdery mildew species [11]. Moreover, a field experiment has clearly demonstrated that genetically differentiated Ampelomyces strains occurring naturally in certain powdery mildew species can easily disperse and parasitize other powdery mildew species on their respective host plants [11]. Furthermore, a broad sampling campaign revealed that grapevine powdery mildew was naturally parasitized by phylogenetically diverse Ampelomyces strains in the field [38].
Some of these previous studies, and the results reported in this work, indicate that Ampelomyces mycoparasites are not strictly specialized on the powdery mildew species in which they are found in the field, with each cluster instead naturally parasitizing a wide range of powdery mildew species. Moreover, this study provides the first evidence for recombination events in Ampelomyces strains isolated from different, non-APM mycohost species in the field. The sexual fruiting bodies of Ampelomyces have never been observed, despite intensive morphological studies on this mycoparasite [37]. The recombination footprints detected may, therefore, be the result of ancient sexual events, before a loss of sex, or recombination may result from a parasexual process in the hyphal anastomoses within the powdery mildew mycelium, the sole habitat of Ampelomyces. It may also be that sex events are very rare and were not observed so far.
Despite their interesting life style and their practical use as commercially available biological control agents for plant pathogenic fungi, very little is known about the population genetics of mycoparasites. This work, together with our earlier study (Kiss et al. 2011), provides the first insight into this domain, based on a reliable set of genetic tools tested with hundreds of strains, and could be used to promote the use of this natural tritrophic relationship between Ampelomyces mycoparasites, their powdery mildew fungal hosts, and the plant hosts of the mildews, as a model system for the study of such interactions. Furthermore, Ampelomyces mycoparasites appear to be among the rare genuine generalist pathogenic fungi, able to parasitize multiple hosts in natural populations. They therefore constitute an excellent model for studying the evolution of pathogens towards a generalist rather than a host-specific strategy [1–3].
Supporting Information
The structure has been inferred by STRUCTURE for K = 2 to K = 7. The STRUCTURE program could form well-defined clusters up to K = 6, indicating the existence of six genetically differentiated clusters. The two different solutions found by Structure at certain K values are shown. The strains are in the same order as in Fig. 1.
(TIF)
(TIF)
The populations are defined based on STRUCTURE assignations (Fig. 1 at K = 6).
(TIF)
(TIF)
The 11 strains used to develop the microsatellite markers are shown in red boldface. Strains of Ampelomyces were designated with upper case letters and/or numbers. When more than one strain was isolated from the same site/plant, these were distinguished by lower case letters (e.g., B119-a and B119-b). If available, public culture collection designations of the strains are also shown. Lower case designations (b1-b365) were applied for apple leaf samples, preserved as herbarium materials and used in the microsatellite genotyping work. Dates and places of collections given with all known details. If several strains were used, collected from more than one site within a locality, or from more than one plant individual within one site, the site and/or the plant number was shown in the table. The identities of the host fungal and host plant species of the strains obtained from earlier works were determined by their suppliers.
(PDF)
(PDF)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Hungarian Scientific Research Fund OTKA NN100415 to LK (http://otka.hu/en) and Janos Bolyai Research Fellowship to PA (http://mta.hu/cikkek/?node_id=26057). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Genetic Marker Services provided support in the form of salaries for author NH, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Barrett LG, Heil M. Unifying concepts and mechanisms in the specificity of plant-enemy interactions. Trends Plant Sci. 2012;17: 282–292. 10.1016/j.tplants.2012.02.009 [DOI] [PubMed] [Google Scholar]
- 2. Bruns E, Carson ML, May G. The Jack of all trades is a master of none: a pathogen's ability to infect a greater number of host genotypes comes at a cost of delayed reproduction. Evolution 2014; 68: 2453–2466. 10.1111/evo.12461 [DOI] [PubMed] [Google Scholar]
- 3. van Tienderen PH. Evolution of generalists and specialists in spatially heterogeneous environments. Evolution 1991;45: 1317–1331. [DOI] [PubMed] [Google Scholar]
- 4. Whitlock MC. The red queen beats the Jack-of-all-trades: the limitations of phenotypic plasticity and niche breadth. Am Nat. 1996;148: S65–S77. [Google Scholar]
- 5. Soler JJ, Martin Vivaldi M, Moller AP. Geographic distribution of suitable hosts explains the evolution of specialized gentes in the European cuckoo Cuculus canorus . BMC Evol Biol. 2009;9: 88 10.1186/1471-2148-9-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rice WR. Disruptive selection on habitat preference and the evolution of reproductive isolation: a simulation study. Evolution 1984;38: 1251–1260. [DOI] [PubMed] [Google Scholar]
- 7. Johnson PA, Hoppensteadt FC, Smith JJ, Bush GL. Conditions for sympatric speciation: a diploid model incorporating habitat fidelity and non-habitat assortative mating. Evol Ecol. 1996;10: 187–205. [Google Scholar]
- 8. Giraud T, Refrégier G, de Vienne DM, Le Gac M, Hood ME. Speciation in fungi Fung Genet Biol. 2008;45: 791–802. 10.1016/j.fgb.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 9. Le Gac M, Giraud T. Existence of a pattern of reproductive character displacement in Basidiomycota but not in Ascomycota. J Evol Biol. 2008;21: 761–772. 10.1111/j.1420-9101.2008.01511.x [DOI] [PubMed] [Google Scholar]
- 10. Théron A, Combes C. Asynchrony of infection timing, habitat preference, and sympatric speciation of schistosome parasites. Evolution 1995;49: 372–375. [DOI] [PubMed] [Google Scholar]
- 11. Kiss L, Pintye A, Kovacs GM, Jankovics T, Fontaine MC, Harvey N, et al. Temporal isolation explains host-related genetic differentiation in a group of widespread mycoparasitic fungi. Mol Ecol. 2011;20: 1492–1507. 10.1111/j.1365-294X.2011.05007.x [DOI] [PubMed] [Google Scholar]
- 12. Giraud T, Villaréal L, Austerlitz F, Le Gac M, Lavigne C. Importance of the life cycle in host race formation and sympatric speciation in parasites. Phytopathology 2006;96: 280–287. 10.1094/PHYTO-96-0280 [DOI] [PubMed] [Google Scholar]
- 13. Giraud T, Gladieux P, Gavrilets S. Linking emergence of fungal plant diseases and ecological speciation. Trends Ecol Evol. 2010;25: 387–395. 10.1016/j.tree.2010.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gladieux P, Guerin F, Giraud T, Caffier V, Lemaire C, Parisi L, et al. Emergence of novel fungal pathogens by ecological speciation: importance of the reduced viability of immigrants. Mol Ecol. 2011;20: 4521–4532. 10.1111/j.1365-294X.2011.05288.x [DOI] [PubMed] [Google Scholar]
- 15. Fournier E, Giraud T. Sympatric genetic differentiation of a generalist pathogenic fungus, Botrytis cinerea, on two different host plants, grapevine and bramble. J Evol Biol. 2008;21: 122–132. [DOI] [PubMed] [Google Scholar]
- 16. Gibson AK, Hood ME, Giraud T. Sibling competition arena: selfing and a competition arena can combine to constitute a barrier to gene flow in sympatry. Evolution 2012;66: 1917–1930. 10.1111/j.1558-5646.2011.01563.x [DOI] [PubMed] [Google Scholar]
- 17. Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, et al. Phylogenetics species recognition and species concepts in Fungi. Fung Genet Biol. 2000;31: 21–32. [DOI] [PubMed] [Google Scholar]
- 18. Le Gac M, Hood ME, Fournier E, Giraud T. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution 2007;61: 15–26. [DOI] [PubMed] [Google Scholar]
- 19. Giraud T, Fortini D, Levis C, Lamarque C, Leroux P, LoBuglio K, et al. Two sibling species of the Botrytis cinerea complex, transposa and vacuma, are found in sympatry on numerous host plants. Phytopathology 1999;89: 967–973. 10.1094/PHYTO.1999.89.10.967 [DOI] [PubMed] [Google Scholar]
- 20. Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea . Plos Genet. 2011;7: e1002230 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farrer RA, Weinert LA, Bielby J, Garner TWJ, Balloux F, Clare F, et al. Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc Natl Acad Sci USA. 2011;108: 18732–18736. 10.1073/pnas.1111915108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012;484: 186–194. 10.1038/nature10947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barres B, Halkett F, Dutech C, Andrieux A, Pinon J, Frey P. Genetic structure of the poplar rust fungus Melampsora larici-populina: Evidence for isolation by distance in Europe and recent founder effects overseas. Infect Genet Evol. 2008;8: 577–587. 10.1016/j.meegid.2008.04.005 [DOI] [PubMed] [Google Scholar]
- 24. Giraud T, Enjalbert J, Fournier E, Delmotte F, Dutech C. Population genetics of fungal diseases of plants. Parasite-Journal De La Societe Francaise De Parasitologie 2008;15: 449–454. [DOI] [PubMed] [Google Scholar]
- 25. Giraud T. Patterns of within population dispersion and mating of the fungus Microbotryum violaceum parasitising the plant Silene latifolia . Heredity 2004;93: 559–565. [DOI] [PubMed] [Google Scholar]
- 26. Grunwald NJ, Hoheisel GA. Hierarchical analysis of diversity, selfing, and genetic differentiation in populations of the oomycete Aphanomyces euteiches . Phytopathology 2006;96: 1134–1141. 10.1094/PHYTO-96-1134 [DOI] [PubMed] [Google Scholar]
- 27. Rieux A, De Bellaire LD, Zapater MF, Ravigne V, Carlier J. Recent range expansion and agricultural landscape heterogeneity have only minimal effect on the spatial genetic structure of the plant pathogenic fungus Mycosphaerella fijiensis . Heredity 2013;110: 29–38. 10.1038/hdy.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rieux A, Halkett F, de Bellaire LD, Zapater MF, Rousset F, Ravigne V, et al. Inferences on pathogenic fungus population structures from microsatellite data: new insights from spatial genetics approaches. Mol Ecol. 2011;20: 1661–1674. 10.1111/j.1365-294X.2011.05053.x [DOI] [PubMed] [Google Scholar]
- 29. Gladieux P, Zhang XG, Afoufa-Bastien D, Sanhueza RMV, Sbaghi M, Le Cam B. On the origin and spread of the scab disease of apple: out of central Asia. PLoS One 2008;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dutech C, Rossi JP, Fabreguettes O, Robin C. Geostatistical genetic analysis for inferring the dispersal pattern of a partially clonal species: example of the chestnut blight fungus. Mol Ecol. 2008;17: 4597–4607. 10.1111/j.1365-294X.2008.03941.x [DOI] [PubMed] [Google Scholar]
- 31. Rieux A, Lenormand T, Carlier J, de Bellaire LD, Ravigne V. Using neutral cline decay to estimate contemporary dispersal: a generic tool and its application to a major crop pathogen. Ecol Lett. 2013;16: 721–730. 10.1111/ele.12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gladieux P, Byrnes E, Fisher M, Aguileta G, Heitman J, Giraud T. Epidemiology and evolution of fungal pathogens, in plants and animals In: T. M, editor editors. Genetics and Evolution of Infectious Diseases. Elsevier; 2010; pp. 59–132. [Google Scholar]
- 33. Millanes A, Truong C, Westberg M, Diederich P, Wedin M. Host switching promotes diversity in host-specialized mycoparasitic fungi: uncoupled evolution in the Biatoropsis-Usnea system. Evolution 2014;68: 1576–1593. 10.1111/evo.12374 [DOI] [PubMed] [Google Scholar]
- 34. Molina M, DePriest P, Lawrey J. Genetic variation in the widespread lichenicolous fungus Marchandiomyces corallinus . Mycologia 2005;97: 454–463. [DOI] [PubMed] [Google Scholar]
- 35. Werth S, Millanes A, Wedin M, Scheidegger C. Lichenicolous fungi show population subdivision by host species but do not share population history with their hosts. Fungal Biology 2013;117: 71–84. 10.1016/j.funbio.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 36. Kiss L, Russell J, Szentivanyi O, Xu X, Jeffries P. Biology and biocontrol potential of Ampelomyces mycoparasites, natural antagonists of powdery mildew fungi. Biocontrol Sci Technol 2004;14: 635–651. [Google Scholar]
- 37. Kiss L. Intracellular mycoparasites in action: interactions between powdery mildew fungi and Ampelomyces In: Avery S., Stratford M. and Van West P., editors. Stress in Yeasts and Filamentous Fungi. London: Academic Press, Elsevier; 2008; pp. 37–52. [Google Scholar]
- 38. Pintye A, Bereczky Z, Kovacs GM, Nagy LG, Xu XM, Legler SE, et al. No Indication of strict host associations in a widespread mycoparasite: grapevine powdery mildew (Erysiphe necator) Is attacked by phylogenetically distant Ampelomyces strains in the field. Phytopathology 2012;102: 707–716. 10.1094/PHYTO-10-11-0270 [DOI] [PubMed] [Google Scholar]
- 39. Park M-J, Choi Y-J, Hong S-B, Shin H-D. Genetic variability and mycohost association of Ampelomyces quisqualis isolates inferred from phylogenetic analyses of ITS rDNA and actin gene sequences. Fungal Biol. 2010;114: 235–247. 10.1016/j.funbio.2010.01.003 [DOI] [PubMed] [Google Scholar]
- 40. Liang C, Yang JR, Kovacs GM, Szentivanyi O, Li BD, Xu XM, et al. Genetic diversity of Ampelomyces mycoparasites isolated from different powdery mildew species in China inferred from analyses of rDNA ITS sequences. Fungal Diversity 2007;24: 225–240. [Google Scholar]
- 41. Szentivanyi O, Kiss L, Russell JC, Kovacs GM, Varga K, Jankovics T, et al. Ampelomyces mycoparasites from apple powdery mildew identified as a distinct group based on single-stranded conformation polymorphism analysis of the rDNA ITS region. Mycol Res. 2005;109: 429–438. [DOI] [PubMed] [Google Scholar]
- 42. Legler SE, Caffi T, Kiss L, Pintye A, Rossi V. Methods for screening new Ampelomyces strains to be used as biocontrol agents against grapevine powdery mildew. IOBC/WPRS Bull. 2011;67: 149–154. [Google Scholar]
- 43. Ropars J, Dupont J, Fontanillas E, de la Vega RCR, Malagnac F, Coton M, et al. Sex in cheese: evidence for sexuality in the fungus Penicillium roqueforti . PLoS One 2012;7: e49665 10.1371/journal.pone.0049665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burt A, Carter DA, Koenig GL, White TJ, Taylor JW. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis . Proc Natl Acad Sci USA. 1996;93: 770–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dyer PS, O'Gorman CM. A fungal sexual revolution: Aspergillus and Penicillium show the way. Curr Op Microbiol. 2011;14: 649–654. [DOI] [PubMed] [Google Scholar]
- 46. Giraud T, Levis C, Fortini D, Leroux P, Brygoo Y. RFLP markers show genetic recombination in Botrytis cinerea and transposable elements reveal two sympatric species. Mol Biol Evol. 1997;14: 1177–1185. [DOI] [PubMed] [Google Scholar]
- 47. Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, et al. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol. 2005;15: 1242–1248. [DOI] [PubMed] [Google Scholar]
- 48. O'Gorman CM, Fuller HT, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus . Nature 2009;457: 471–U475. 10.1038/nature07528 [DOI] [PubMed] [Google Scholar]
- 49. Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbio Mol Biol Rev. 2010;74: 298–340. 10.1128/MMBR.00005-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hoff B, Poeggeler S, Kueck U. Eighty years after its discovery, Fleming's Penicillium strain discloses the secret of its sex. Euk Cell 2008;7: 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lopez-Villavicencio M, Aguileta G, Giraud T, de Vienne DM, Lacoste S, Couloux A, et al. Sex in Penicillium: Combined phylogenetic and experimental approaches. Fung Genet Biol. 2010;47: 693–706. 10.1016/j.fgb.2010.05.002 [DOI] [PubMed] [Google Scholar]
- 52. Harvey N. Characterization of six polymorphic microsatellite loci from Ampelomyces quisqualis, intracellular mycoparasite and biocontrol agent of powdery mildew. Mol Ecol Notes. 2006;6: 1188–1190. [Google Scholar]
- 53. Kiss L, Nakasone KK. Ribosomal DNA internal transcribed spacer sequences do not support the species status of Ampelomyces quisqualis, a hyperparasite of powdery mildew fungi. Curr Genet. 1998;33: 362–367. [DOI] [PubMed] [Google Scholar]
- 54. Sullivan RF, White JF. Phoma glomerata as a mycoparasite of powdery mildew. App Env Microbiol. 2000;66: 425–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Falk SP, Gadoury DM, Pearson RC, Seem RC. Partial central of grape powdery mildew by the mycoparasite Ampelomyces quisqualis . Plant Dis. 1995;79: 483–490. [Google Scholar]
- 56. Takamatsu S. Phylogeny and evolution of the powdery mildew fungi (Erysiphales, Ascomycota) inferred from nuclear ribosomal DNA sequences. Mycoscience 2004;45: 147–157. [Google Scholar]
- 57. Huson D, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23: 254–267. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16221896. [DOI] [PubMed] [Google Scholar]
- 58.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. 1996–2004.
- 59. Raymond M, Rousset F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Hered. 1995;86:: 248–249. [Google Scholar]
- 60. Rousset F. Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Res. 2008;8: 103–106. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21585727. 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 61. Arnaud-Haond S, Belkhir K. GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes 2007;7: 15–17. [Google Scholar]
- 62. Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000;155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jakobsson M, Rosenberg N. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007;23: 1801–1806. [DOI] [PubMed] [Google Scholar]
- 64. Rosenberg N. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4: 137–138. [Google Scholar]
- 65. Earl D, VonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res. 2012;4: 359–361. [Google Scholar]
- 66. Pritchard J, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000;155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- 68.Angeli D, Pellegrini E, Micheli S, Maurhofer M, Gessler C, Pertot I. Is the biocontrol efficacy of Ampelomyces quisqualis against powdery mildew related to the aggressiveness of the strain?. In: A. Calonnec, editor. Proc 6th Int Workshop on Grapevine Downy and Powdery Mildew. Bordeaux, France: INRA ISVV. 2010; pp. 164–166.
- 69. Sztejnberg A, Galper S, Mazar S, Lisker N. Ampelomyces quisqualis for biological and integrated control of powdery mildex in Israel. J Phytopathol. 1989;124: 285–295. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The structure has been inferred by STRUCTURE for K = 2 to K = 7. The STRUCTURE program could form well-defined clusters up to K = 6, indicating the existence of six genetically differentiated clusters. The two different solutions found by Structure at certain K values are shown. The strains are in the same order as in Fig. 1.
(TIF)
(TIF)
The populations are defined based on STRUCTURE assignations (Fig. 1 at K = 6).
(TIF)
(TIF)
The 11 strains used to develop the microsatellite markers are shown in red boldface. Strains of Ampelomyces were designated with upper case letters and/or numbers. When more than one strain was isolated from the same site/plant, these were distinguished by lower case letters (e.g., B119-a and B119-b). If available, public culture collection designations of the strains are also shown. Lower case designations (b1-b365) were applied for apple leaf samples, preserved as herbarium materials and used in the microsatellite genotyping work. Dates and places of collections given with all known details. If several strains were used, collected from more than one site within a locality, or from more than one plant individual within one site, the site and/or the plant number was shown in the table. The identities of the host fungal and host plant species of the strains obtained from earlier works were determined by their suppliers.
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.