ABSTRACT
CodY is a branched-chain amino acid-responsive transcriptional regulator that controls the expression of several dozen transcription units in Bacillus subtilis. The presence of isoleucine, valine, and leucine in the growth medium is essential for achieving high activity of CodY and for efficient regulation of the target genes. We identified three permeases—BcaP, BraB, and BrnQ—that are responsible for the bulk of isoleucine and valine uptake and are also involved in leucine uptake. At least one more permease is capable of efficient leucine uptake, as well as low-affinity transport of isoleucine and valine. The lack of the first three permeases strongly reduced activity of CodY in an amino acid-containing growth medium. BcaP appears to be the most efficient isoleucine and valine permease responsible for their utilization as nitrogen sources. The previously described strong CodY-mediated repression of BcaP provides a mechanism for fine-tuning CodY activity by reducing the availability of amino acids and for delaying the utilization of isoleucine and valine as nitrogen and carbon sources under conditions of nutrient excess.
IMPORTANCE Bacillus subtilis CodY is a global transcriptional regulator that is activated by branched-chain amino acids (BCAA). Since the level of BCAA achieved by intracellular synthesis is insufficient to fully activate CodY, transport of BCAA from the environment is critical for CodY activation, but the permeases needed for such activation have not been previously identified. This study identifies three such permeases, reports their amino acid transport specificity, and reveals their impact on CodY activation.
INTRODUCTION
CodY is a global transcriptional regulator in Bacillus subtilis that controls, directly or indirectly, the expression of about 200 genes, most of them negatively (1–3). Many of the CodY-regulated genes are involved in nitrogen or carbon metabolism (1, 2, 4–7). CodY homologs are present in most low-G+C Gram-positive bacteria and in many species have been shown to play a global role in metabolic regulation similar to that in B. subtilis, as well as in coordinating expression of virulence-associated functions with expression of metabolic genes (7, 8; see also, e.g., references 9 and 10 and references therein).
B. subtilis CodY is a dimeric 259-residue protein that uses a winged helix-turn-helix motif to bind to DNA (11, 12). The DNA-binding activity of CodY is increased by interaction with two types of effectors, branched-chain amino acids (BCAA; isoleucine [Ile], leucine [Leu], and valine [Val], collectively abbreviated as ILV) (13–17) and GTP (1, 15, 18–20). The effect of GTP on CodY, however, may depend on simultaneous or prior interaction of CodY with ILV because mutant variants of CodY deficient in ILV binding lose most of the ability to regulate CodY-dependent genes (21, 22). The pools of ILV and GTP apparently reflect the nutritional status of the cells, allowing bacteria to change the pattern of CodY-dependent gene expression in response to availability of nutrients in the growth medium.
While the endogenous pool of CodY effectors in B. subtilis cells growing in minimal media is sufficient to regulate some genes (23), the highest activity of CodY is observed in the presence in the medium of ILV and other amino acids (15, 23–27). Thus, the activity of ILV permeases and efficiency of ILV uptake should be critical for activation of CodY.
The identity of ILV permeases in B. subtilis cells has not been established experimentally. The results reported here indicate that three permeases, BcaP, BraB, and BrnQ, are involved in the high-affinity uptake of Ile and Val. At least one more permease is involved in the uptake of Leu, as well as the low-affinity transport of Ile and Val. Strong CodY-mediated repression of BcaP creates a mechanism for fine-tuning of CodY activity.
MATERIALS AND METHODS
Bacterial strains and culture media.
The B. subtilis strains constructed and used in the present study were all derivatives of strain SMY (28) and are described in Table 1 or below; the latter strains were constructed by transformation using chromosomal DNA, isolated from the listed strains. Escherichia coli strain JM107 (29) was used for construction and isolation of plasmids. Cell growth in 0.5% glucose–0.2% ammonium minimal TSS medium and rich DS medium was as described previously (22).
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype or description | Source and/or referencea |
|---|---|---|
| SMY | Prototroph | 28 |
| CU4138 | leuB84::Tn917 trpC2 | 37* |
| CU4139 | liv-1-82::Tn917 trpC2 | 37* |
| QB943 KIT-4 | ilvA1 pyrD1 thyA1 thyB1 trpC2 | 53* |
| PS251 | codY::(erm::spc) trpC2 | P. Serror |
| BB274 | ΔazlB2 (in frame) | 31 |
| BB284 | Δ(azlCD brnQ)::neo | 31 |
| BB2505 | ΔamyE::[erm Φ(bcaP283-lacZ)] lacA::tet | 22 |
| BB2511 | ΔamyE::spc lacA::tet | 27 |
| BB2676 | ΔamyE::[erm Φ(dppA-lacZ)] lacA::tet | 27 |
| BB2726 | ΔbcaP::spc | SMY × pBB1488 |
| BB2770 | ΔamyE::[erm Φ(ybgE292-lacZ)] lacA::tet | 23 |
| BB3046 | ΔbraB::cat | SMY × pBB1586 |
| BB3088 | ΔazlCD | SMY × pBB1595 |
| BB3125 | ΔamyE::[erm Φ(ptb-lacZ)] lacA::tet | BB2511 × pBB1598 |
| BB3165 | bcaPp14 | SMY × pBB1605 |
| BB3198 | bcaPp10/14 | BB3165 × pBB1605 |
*, strains obtained from the Bacillus Genetic Stock Center. × indicates transformation by plasmid DNA.
If indicated, the TSS medium was supplemented with a mixture of 16 amino acids (aa) (24). This mixture contained all amino acids commonly found in proteins except for glutamine, asparagine, histidine, and tyrosine; the concentrations of Ile, Leu, and Val were 200 μg/ml, each. In some experiments Ile, Leu, and Val were omitted from the 16-aa mixture or added separately to TSS. In the latter case, their concentration varied from 20 to 400 μg/ml each. The concentration of Ile or Val was increased to 1 or 2 mg/ml when they it served as the sole nitrogen source in TSS; ammonium was omitted in these experiments.
The following antibiotics were used when appropriate: tetracycline (15 μg/ml), spectinomycin (50 μg/ml), chloramphenicol (2.5 μg/ml), neomycin (2.5 μg/ml), or a combination of erythromycin (0.5 μg/ml) and lincomycin (12.5 μg/ml) for B. subtilis strains and ampicillin (50 μg/ml) for E. coli strains.
DNA manipulations.
Methods for common DNA manipulations, E. coli electroporation, isolation of B. subtilis chromosomal DNA, transformation of B. subtilis cells, and sequence analysis were as previously described (27, 30). All of the oligonucleotides used in the present study are listed in Table 2. Chromosomal DNA of B. subtilis strain SMY was used as the template for PCR. All cloned PCR-generated fragments were verified by sequencing at the Tufts University Core Facility.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′–3′)a | Specificity |
|---|---|---|
| Forward | ||
| oBB244 | GACAGGGATCCATTCATGTGAATGG | bcaP |
| oBB330 | AAAAAGAATTCACTAGTAACTGCAGTGCATCCAAAGCAC | bcaP |
| oBB396 | CCATCTCTAGAACAGGAACTGTTTC | braB |
| oBB426 | TACAATCTAGATAAATATGGCCTTG | ptb |
| Reverse | ||
| oBB245 | TCATTAAGCTTCAACTCCCGATC | bcaP |
| oBB397 | ATAACGGTACCGTTCTGTGACATTAAC | braB |
| oBB296 | CTGTAAAGCTTCTCTCTTTATCAAAAGG | bcaP |
| oBB427 | GTTACAAGCTTTCTTGTTTCGACTC | ptb |
Restriction sites are underlined.
Construction of braB- and bcaP-null mutants.
A 1.07-kb PCR fragment containing most of the braB gene was synthesized using oBB396 and oBB397 as primers and cloned between the XbaI and KpnI sites of an integrative plasmid pBB544 (neo) (31) to create pBB1571. A deletion-insertion mutation within the braB gene was created by replacing the 0.48-kb MfeI-ClaI fragment of pBB1571 with a 1.5-kb EcoRI-ClaI cat cassette, excised from pJPM12 (32). The orientation of the cat gene in the resulting plasmid, pBB1586, coincides with that of the braB gene.
Plasmid pBB1484, containing the 5′ regulatory region and the 3′ end of the bcaP gene, together with the flanking nucleotides, was constructed by ligating two corresponding 0.24- and 0.39-kb PCR products between the BamHI and HindIII sites of an integrative plasmid pJPM1 (cat) (32). The PCR products were synthesized using oBB244 and oBB245 or oBB330 and oBB296 as primers and digested with BamHI and EcoRI (an internal site) or EcoRI and HindIII, respectively. A deletion-insertion mutation within the bcaP gene was created by cloning a 1.18-kb SpeI-PstI spc cassette, excised from pJL73 (33), between the SpeI and PstI sites (both originating from oBB330) of pBB1484. The orientation of the spc gene in the resulting plasmid pBB1488 coincides with that of the bcaP gene.
pBB1586 (braB::cat neo) and pBB1488 (bcaP::spc cat) were introduced into B. subtilis SMY, and Catr Neos or Spcr Cats transformants, arising from double-crossover, homologous recombination events, were selected, respectively. The replacement of the chromosomal wild-type allele by the mutant allele was confirmed by analyzing the size of the chromosomal braB or bcaP allele by PCR.
Construction of an ΔazlCD strain.
The pBB1595 plasmid containing a deletion of the 3′ end of the azlC gene and 5′ end of the azlD gene was constructed after excising, using BsaAI and BsrGI, a 0.35-bp fragment from an integrative plasmid pBB419 (azlBCDE′ cat) (31), blunting the ends, and self-ligating the remaining fragment. The deletion removed 31 and 36% of azlC and azlD, respectively, and fused the remaining coding parts of the genes as an in-frame construct.
pBB1595 (ΔazlCD cat) was introduced by a single-crossover, homologous recombination event into the azl chromosomal locus of strain SMY. Spontaneous Cats colonies indicating excision of pBB1595 from the chromosome were screened for, and colonies acquiring the azlCD deletion were found by analyzing the size of the chromosomal azlCD allele by PCR.
Construction of a bcaPp10/14 strain with the derepressed bcaP gene.
The combination of two regulatory mutations, bcaPp2 and bcaPp14 was shown to virtually abolish CodY-dependent repression of the bcaP gene (22). The PCR fragment that was used to create the 283-bp bcaPp2/14 regulatory region (22) was blunt ended with the DNA polymerase I Klenow fragment, digested with BamHI and cloned between the SmaI and BamHI sites of an integrative plasmid pBB1579 (bgaB neo) (34). Sequencing of the resulting plasmid pBB1605 showed that the plasmid acquired an additional mutation in CodY-binding motif I, a T-to-A transition immediately downstream of the p2 mutation in the central, less conserved nucleotide of the motif (22). As a result, the original sequence of the motif, ATTTTTCTAACAATT, changed to ATTTTTaaAACAATT. The new double mutation in motif I was assigned the allele number p10 and shown, in separate experiments, to have a phenotype identical to that of the p2 mutation (data not shown). pBB1605 (bcaPp10/14 bgaB neo) was introduced by a single-crossover, homologous recombination event into the bcaP chromosomal locus of strain SMY. White Neos colonies indicating excision of pBB1605 from the chromosome were searched for on plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), the colored substrate of bgaB-encoded β-galactosidase. A colony that had acquired the bcaPp14 mutation but not the bcaPp10 double mutation was found by sequencing the PCR product from the chromosomal bcaP allele. The resulting strain, BB3165 (bcaPp14), was subjected to another round of transformation with pBB1605, and strain BB3198 (bcaPp10/14) was isolated as described above.
Construction of a transcriptional ptb-lacZ fusion.
Plasmid pBB1598 (ptb-lacZ erm) was created by cloning a 0.15-kb XbaI- and HindIII-treated ptb PCR product that contains the entire regulatory region of the gene into an integrative plasmid pHK23 (erm) (27). The ptb PCR product was synthesized using oBB426 and oBB427 as primers.
A B. subtilis strain carrying the ptb-lacZ fusion at the amyE locus was isolated after transforming strain BB2511 (amyE::spc lacA) with pBB1598, by selecting for resistance to erythromycin, and screening for the loss of the spectinomycin-resistance phenotype, which indicated a double-crossover, homologous recombination event. Strain BB2511 and its derivatives has very low endogenous β-galactosidase activity due to a null mutation in the lacA gene (35).
Ile uptake.
Cells were grown at 37°C in TSS glucose-ammonium medium until mid-exponential phase, collected under vacuum on 0.45 μM nitrocellulose filters, washed, and resuspended at optical density at 600 nm (OD600) of ∼0.8 in the same medium without NH4Cl but with 100 μg of chloramphenicol/ml to completely prevent incorporation of amino acids into proteins. Further incubation of cells was at 26 or 37°C; the lower temperature was used in order to reduce the high rate of Ile uptake. [14C]Ile (Moravek Biochemicals) was added to 1-ml cultures to 0.1 μCi/ml (10.3 μM), and 160-μl samples were taken at the indicated times, collected immediately under vacuum on 0.45-μm-pore-size nitrocellulose filters, washed with 5 ml of TSS without NH4Cl but containing 76 μM unlabeled Ile, dried, and counted using Ecoscint H scintillation liquid (National Diagnostics). Competing amino acids, if present, were added simultaneously with [14C]Ile at a concentration of 1 mM (100-fold excess). Protein concentration was determined in sonicated cell samples using the Bio-Rad protein assay reagent. One-milliliter culture samples at an OD600 of 1 contained 127.5 μg of total protein. The parameters of the Michaelis-Menten kinetics, Km and Vmax, were determined by a nonlinear regression analysis using the Solver function of Excel. The Ile concentration was varied from 0.8 to 160 μM; the rates of Ile uptake were assumed to be equal to the initial rates of uptake during the first 20 s of the assay.
Enzyme assays.
The β-galactosidase activity was determined as described previously (30).
RESULTS
Phenotypes of brnQ- and braB-null mutants.
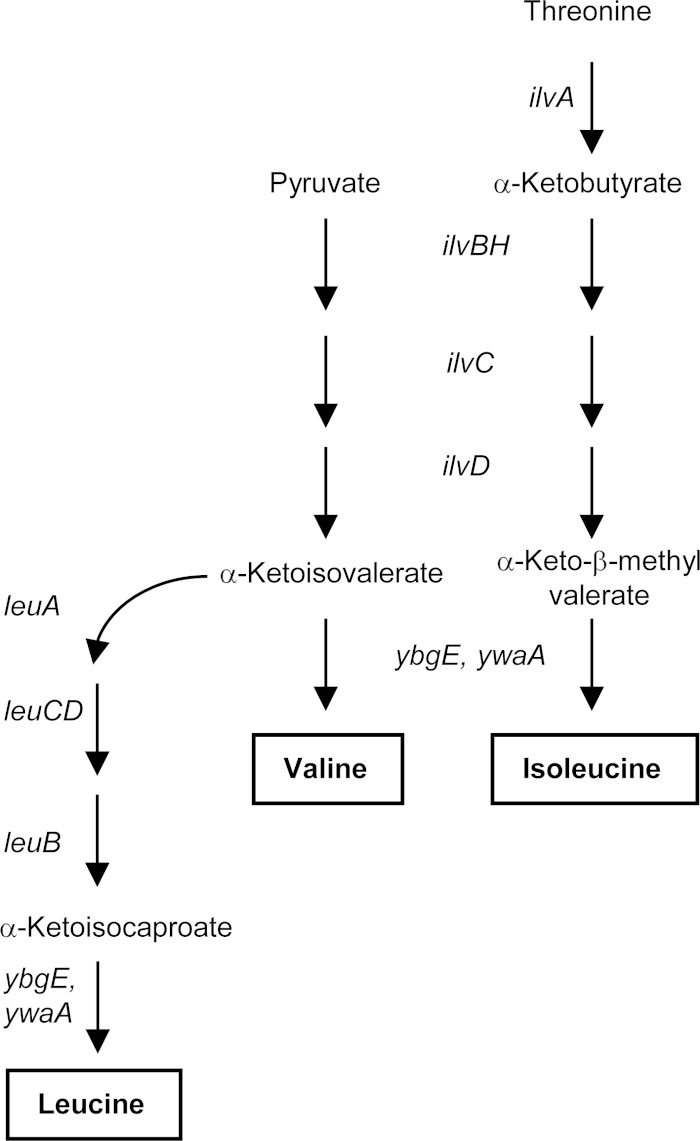
Two B. subtilis genes, brnQ and braB, encode proteins with high similarity to BCAA permeases of other organisms (36). The brnQ (azlE) gene is part of the azlBCDEF operon, which is also involved in export of a leucine analog, 4-azaleucine (31). A deletion of brnQ did not confer any obvious growth defect on B. subtilis liv cells auxotrophic for ILV (the liv-1-82::Tn917 mutation is an insertion within the ilvBHC leuABCD operon that also led to a partial deletion of the operon [37]; see Fig. 1 for the pathway of ILV biosynthesis), i.e., such cells were able to transport enough ILV from an ILV-containing (20 or 40 μg/ml [0.15 or 0.3 mM] each) TSS glucose-ammonium medium to grow at the same rate as wild-type cells (31). Since a mutation in the transporter solely responsible for the uptake of at least one of the BCAAs would prevent growth of an auxotroph in this medium, it was concluded that BrnQ is not the only BCAA permease under these growth conditions.
FIG 1.
Pathway of ILV biosynthesis in B. subtilis.
A deletion-insertion in the braB gene was constructed as described in Materials and Methods. Double or triple null mutant strains BB3465 (braB liv) and BB3065 (braB brnQ liv) were still able to take up ILV to fully satisfy their growth requirements and grew as well as a single liv mutant, strain BB3467 (Fig. 2 and data not shown), indicating that neither BraB alone nor the combination of BraB and BrnQ serve as the only BCAA permease(s) under these conditions. Thus, at least one additional permease must be capable of transporting ILV under these growth conditions.
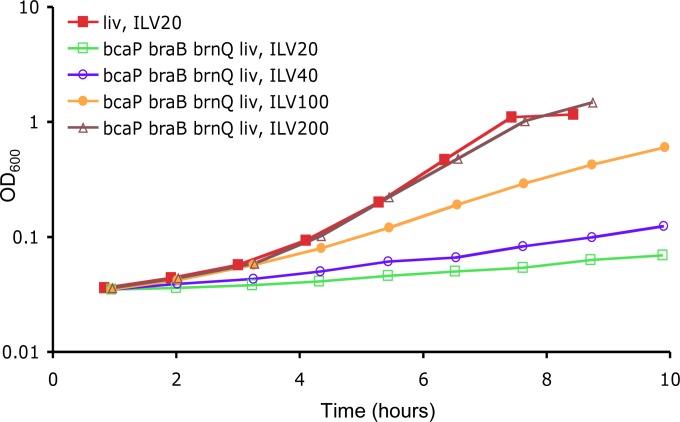
FIG 2.
Growth of auxotrophic liv mutant strains with different concentrations of ILV. Cells of strain BB3467 (liv) or BB3067 (bcaP braB brnQ liv) were grown overnight in TSS glucose-ammonium minimal medium with 200 μg of ILV/ml and then diluted 100-fold in the same medium containing various concentrations of ILV, as indicated. Each growth experiment was performed at least twice, and the results of a representative experiment are shown.
Phenotype of a bcaP-null mutant.
The BcaP protein (also known as CtrA) was described as a major transporter of BCAAs in Lactococcus lactis (38). This protein is only moderately similar (34% identity) to B. subtilis YhdG. However, both the L. lactis bcaP and the B. subtilis yhdG genes are among the most highly CodY-regulated genes in each organism, suggesting that they may have similar functions (1, 22, 39). Considering the role of YhdG in BCAA transport, described below, and its similarity to L. lactis BcaP, we have renamed the yhdG gene as bcaP (22).
A deletion-insertion in the B. subtilis bcaP gene was constructed as described in Materials and Methods. In the TSS medium supplemented with ILV (20 μg/ml each), the ILV auxotrophic liv strain containing the bcaP-null mutation (BB2738) grew as well as the single liv mutant (data not shown). Triple null mutant strains BB3057 (bcaP braB liv) and BB2917 (bcaP brnQ liv) also had little or no growth defect in the presence of ILV, similar to the braB brnQ liv mutant described above (doubling times for all triple mutant strains and for the single liv mutant strain varied between 0.79 and 0.85 h). However, the growth rate of the quadruple null mutant strain BB3067 (braB brnQ bcaP liv) was reduced dramatically (doubling time of 4 to 7 h) (Fig. 2).
Rather unexpectedly, even the quadruple mutant was able to grow at almost normal rate (doubling time ∼1 h) in the minimal medium supplemented with higher concentrations of ILV (200 μg/ml each) (Fig. 2). As expected, no growth defect was observed for the prototrophic (liv+) triple null mutant braB brnQ bcaP (strain BB3051, data not shown). All mutants described above were able to grow on rich DSM agar plates, although some of them formed colonies of reduced size; the quadruple null mutant BB3067 had the largest growth defect (data not shown).
We conclude that BcaP, BraB, and BrnQ all contribute and together are required for the efficient, high-affinity uptake of at least one of the three BCAAs, indicating that each of these proteins is a BCAA permease. Moreover, the individual activities of each of the three permeases are sufficient for providing enough BCAAs for the unimpeded growth of the auxotrophic liv strain with moderate concentrations of ILV. We also conclude that at least one additional permease with low affinity for BCAA is present in B. subtilis cells and can supply ILV for an auxotrophic strain if an excess of these amino acids is present in the medium.
Uptake of individual branched-chain amino acids.
Using the liv mutant, which is auxotrophic for all three BCAA, it is difficult to determine the extent to which BcaP, BraB, and BrnQ are involved in the transport of individual amino acids. To address this question, we introduced null mutations in the BCAA permease genes into ilvA or leuB mutant strains, auxotrophic only for Ile or Leu, respectively (Fig. 1). For the derivatives of the ilvA mutant, the results matched those for the derivatives of the liv mutant, i.e., the cells failed to grow in the Ile-containing minimal medium (40 μg/ml) only if all three permeases were inactivated; doubling times for all other strains were ∼0.8 h. Surprisingly, no growth defect was observed for the leuB strain in the Leu-containing minimal medium (40 μg/ml) even in the absence of all three of the permeases; as expected, the strain was unable to grow without Leu (data not shown). Thus, in contrast to the situation with Ile, at least one more permease acts as a self-sufficient, high-affinity transporter for Leu, and the roles of BcaP, BraB, and BrnQ in Leu transport remain to be established (see below).
We conclude that the growth defects of the liv derivatives described above are due to the roles of BcaP, BraB, and BrnQ in Ile (and possibly Val) transport. The roles of these permeases in Val transport cannot be assessed by the genetic approach described above as Val auxotrophy is always accompanied by Ile auxotrophy due to shared enzymes in the pathways of Ile and Val biosynthesis (Fig. 1) (40).
Utilization of BCAAs as the sole nitrogen source.
B. subtilis cells are able to utilize Ile or Val but not Leu as the sole nitrogen source in glucose minimal medium (41, 42). The cells of the braB brnQ double null mutant (strain BB3050) were able to utilize either Ile or Val as the sole nitrogen source as efficiently as wild-type cells (doubling time of about 2 h for either amino acid) (Table 3). However, the bcaP-null mutant (strain BB2726) and the bcaP braB brnQ triple null mutant (strain BB3051) lost the ability to utilize either amino acid as the sole nitrogen source (Table 3). Because neither BraB nor BrnQ nor any other transporter was able to compensate for the loss of BcaP in these experiments, BcaP is the only permease in wild-type cells that allows the efficient uptake of the massive amount of Ile or Val required for its utilization as the sole nitrogen source.
TABLE 3.
Growth of B. subtilis strains with Ile or Val as the sole nitrogen sourcea
| Strain | Relevant genotype | Doubling time (h) |
|
|---|---|---|---|
| Ile | Val | ||
| SMY | Wild type | 1.87 | 1.92 |
| BB3050 | braB brnQ | 1.81 | 1.89 |
| BB2726 | bcaP | NGb | NG |
| BB3051 | bcaP braB brnQ | NG | NG |
| BB3085 | azlB bcaP braB | 1.82 | 2.00 |
Cells were grown in TSS glucose minimal medium, in which 0.1% Ile or 0.1% Val was substituted for 0.2% ammonium chloride as the sole nitrogen source.
NG, no growth.
In our previous work, we showed that inactivation of the azlB gene, encoding a repressor of the azlBCDEF operon, leads to a significant overexpression of downstream genes, including the brnQ (azlE) gene (31). In accord with this result, the azlB bcaP braB triple mutant (strain BB3085) regained ability to utilize both Ile and Val as sole nitrogen sources despite the absence of BcaP (Table 3). These results indicate that BrnQ-mediated BCAA uptake is limited by the low expression of the brnQ gene and that BrnQ, as well as BcaP, is capable of transporting both Ile and Val.
Expression of the ptb operon as a test for ILV uptake.
The B. subtilis ptb bcd buk lpdV bkdAA bkdAB bkdB operon is involved in the synthesis of branched-chain fatty acids and degradation of BCAA and was shown to be induced by addition of Ile or Val (42). The ability of these two amino acids to induce the ptb-lacZ fusion was reduced but not completely abolished in the bcaP braB brnQ triple null mutant (Table 4, strain BB3143). Thus, as indicated above, B. subtilis cells have an additional permease(s) with a specificity for Ile and Val.
TABLE 4.
Role of ILV uptake in the activation of the ptb-lacZ fusiona
| Strain | Relevant genotype | Addition(s) to the medium | β-Galactosidase activity (Miller units) |
|---|---|---|---|
| BB3125 | Wild type | None | 0.22 |
| Ile | 56.4 | ||
| Val | 40.5 | ||
| Leu | 2.44 | ||
| Ile + 13 aa | 12.9 | ||
| BB3143 | bcaP braB brnQ | None | 0.22 |
| Ile | 10.2 | ||
| Val | 3.32 | ||
| Leu | 1.07 | ||
| Ile + 13 aa | 0.80 | ||
| BB3246 | bcaP brnQ | Ile | 35.8 |
| Val | 12.5 | ||
| BB3250 | braB brnQ | Ile | 44.7 |
| Val | 16.4 | ||
| BB3290 | bcaP braB | Ile | 25.9 |
| Val | 8.78 | ||
| BB3291 | azlB2 bcaP braB | Ile | 108.4 |
| Val | 46.9 | ||
| Ile + 13 aa | 36.5 | ||
| BB3285 | bcaPp10/14 | Ile | 114.0 |
| Val | 68.8 | ||
| Ile + 13 aa | 21.3 |
Cells were grown in TSS glucose-ammonium medium with or without Ile, Leu, or Val (200 μg/ml) and a mixture of 13 amino acids. The β-galactosidase specific activity was assayed and expressed in Miller units. All values are averages from at least two experiments, and the mean errors did not exceed 30%.
Expression from the ptb promoter was also reduced in each of the double permease mutants, indicating that none of the three identified permeases, even with the help of a yet unknown permease(s), was able to provide enough Ile or Val for full activation of ptb, despite their high concentration (200 μg/ml) in the medium (Table 4). The role of BraB in ptb expression, observed by comparing bcaP braB brnQ and bcaP brnQ mutants, shows that BraB, as well as BcaP and BrnQ, is able to transport both Ile and Val. In contrast to Ile and Val, Leu was able to cause much weaker activation of the ptb promoter (Table 4, strain BB3125). This weak effect was further reduced in the bcaP braB brnQ triple null mutant, indicating that at least one of the corresponding permeases is involved in Leu uptake (Table 4, strain BB3143).
The ability of Ile to induce ptb-lacZ expression was significantly reduced by the presence of a mixture of 13 amino acids (Table 4). This indicates that Ile-transporting activities of several BCAA permeases, including the low-affinity one, which is active in the bcaP braB brnQ triple null mutant, are likely reduced under these conditions either due to competition or inhibition by other amino acids or to lower expression of the corresponding genes. In contrast, derepression of BrnQ due to the azlB-null mutation led to the higher induction of the ptb-lacZ fusion in the presence of either Ile or Val (Table 4, strain BB3291). Similarly, derepression of bcaP due to the presence of the p10/p14 mutation (see below), which makes the gene independent of CodY, also led to higher induction of the ptb-lacZ fusion (Table 4, strain BB3285).
Isoleucine uptake assays.
Resting wild-type B. subtilis cells were able to efficiently take up Ile (Fig. 3A). Note that our uptake measurements did not preclude Ile metabolism; however, because Ile incorporation into proteins was prevented by the lack of a nitrogen source in the assay medium and the addition of chloramphenicol, Ile uptake, measured in our experiments, mostly reflected Ile transport. Inactivation of any two of three BCAA permeases, identified in our work, led to a reduction in the rate of Ile uptake (Fig. 3A). BcaP appeared to be the most efficient Ile permease when present alone, followed by BraB. BrnQ had very low activity in Ile uptake; however, as shown above, even this low activity was sufficient to provide enough Ile for unimpeded growth of an auxotrophic strain. In the bcaP braB brnQ triple null mutant strain, Ile uptake was reduced to an extremely low, but still detectable, level (Fig. 3A). Thus, BcaP, BraB, and BrnQ are responsible for the bulk of Ile uptake, although, as other experiments indicated, another low-efficiency Ile permease(s) exists in B. subtilis cells.
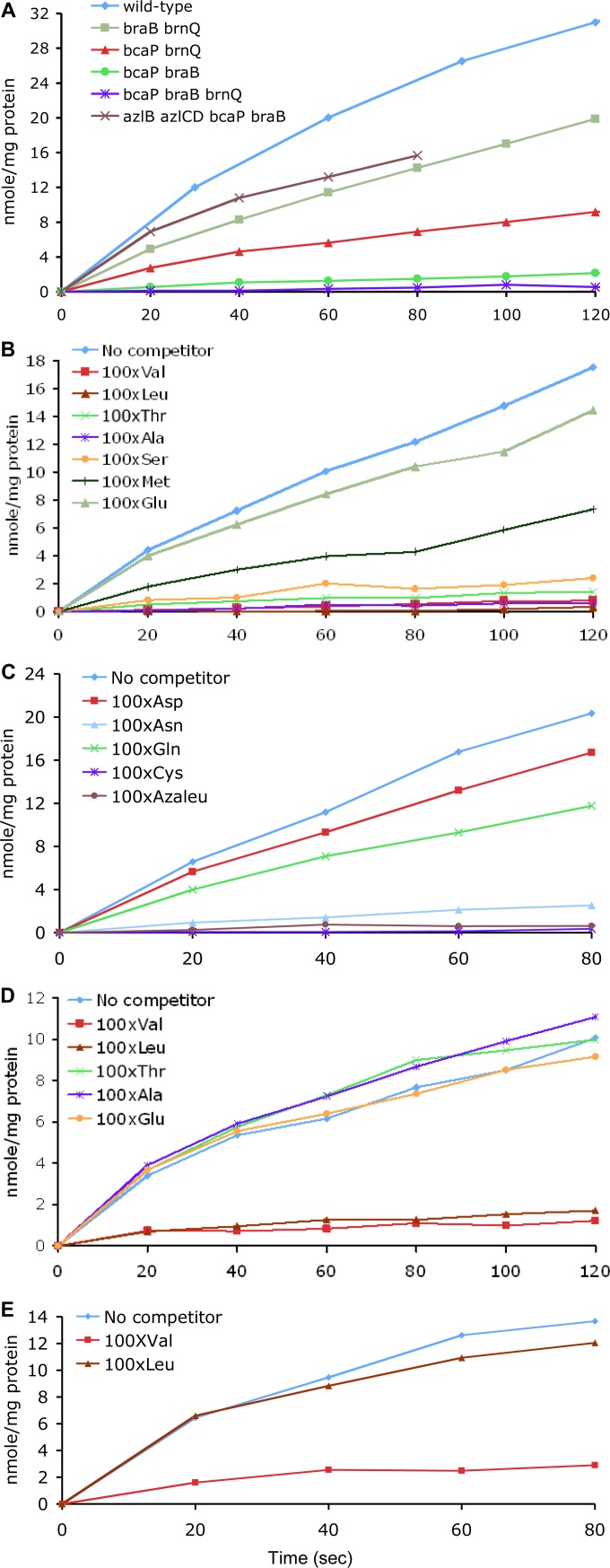
FIG 3.
Uptake of [14C]Ile. Cells were grown in TSS glucose-ammonium minimal medium, and uptake of [14C]Ile was measured as described in Materials and Methods at 26°C (A, B, C, and E) or 37°C (D). (A) SMY (wild-type), BB3050 (braB brnQ), BB2913 (bcaP brnQ), BB3055 (bcaP braB), BB3051 (bcaP braB brnQ), and BB3475 (azlB azlCD braB bcaP); (B and C) BB3050 (braB brnQ); (D) BB2913 (bcaP brnQ); (E) BB3475 (azlB azlCD braB bcaP). A 100-fold excess (1 mM) of competing amino acids was added, as indicated, in panels B to E.
By varying the Ile concentration in the uptake assays, we found that BcaP had the highest affinity for Ile with a Km ≈ 4.1 μM; the Vmax at 26°C was ∼15 nmol (min mg of protein)−1. BraB had a lower affinity for Ile [Km ≈ 16 μM and Vmax ≈ 25 nmol (min mg of protein)−1 at 37°C]. The activity of BrnQ was too low for kinetic determinations in a strain with intact AzlB-mediated regulation of the brnQ gene. However, much higher activity of BrnQ was detected in an unregulated azlB azlCD bcaP braB mutant (Fig. 3A) (see below for the description of the azlCD mutation); a Km of ∼17 μM and a Vmax of ∼59 nmol (min mg of protein)−1 were determined with derepressed BrnQ at 37°C. Ile uptake provided by BcaP was efficiently reduced in the presence of 1 mM (i.e., 100-fold excess over Ile) Val, Leu, alanine, threonine, serine, cysteine, asparagine, and a nonproteinaceous amino acid 4-azaleucine, indicating that either BcaP is able to transport these amino acids or its activity is inhibited in the presence of these amino acids. No other proteinaceous amino acid at 100-fold excess affected BcaP activity by >2-fold (Fig. 3B and C and data not shown). BraB activity was affected by Val and Leu but not by any other proteinaceous amino acid, indicating that BraB is a dedicated BCAA transporter (Fig. 3D and data not shown). The activity of BrnQ in the azlB azlCD bcaP braB strain was significantly affected only by Val (Fig. 3E and data not shown). Thus, BrnQ appears not to be involved in Leu transport. The analysis of the full specificity spectra of BcaP, BraB, and BrnQ and their affinities for various amino acids was beyond the scope of the present study.
Role of ILV uptake and BCAA permeases in the regulation of CodY activity.
By synthesizing amino acids intracellularly, B. subtilis cells produce a level of ILV sufficient to enable protein biosynthesis and other metabolic processes. These concentrations of ILV also support a low level of CodY activity that is sufficient for partial regulation of some genes, such as ybgE, but not others (23). Mutations that increase expression of the ILV biosynthetic pathway can lead to increased ILV pools and higher-level activation of CodY (43). However, the maximal level of CodY activity is observed only in the presence of exogenous, ILV-containing amino acid mixtures (15, 24–27). Under such conditions, i.e., in the glucose-ammonium medium containing ILV (200 μg/ml each) and 13 other amino acids, referred to here as the 16-aa-containing medium, endogenous ILV synthesis does not contribute to CodY activity, because the repression level of several highly CodY-regulated promoters is identical in wild-type cells and auxotrophic liv mutants unable to synthesize ILV (Table 5). A similar effect was observed in the presence of ILV only (Table 5). Thus, uptake of exogenous ILV is absolutely critical for attaining high CodY activity and the efficiency of such uptake is likely to determine the level of CodY activation.
TABLE 5.
Role of intracellular ILV synthesis and BCAA permeases in CodY activitya
| Strain | Relevant genotype | Fusion type | β-Galactosidase activity (Miller units) |
||
|---|---|---|---|---|---|
| No additions | ILV | ILV + 13 aa | |||
| BB2676 | Wild type | dppA-lacZ | 90.3 | 37.3 | 5.79 |
| BB3152 | liv | NG | 36.2 | 4.56 | |
| BB2505 | Wild type | bcaP-lacZ | 55.9 | 7.47 | 0.14 |
| BB3150 | liv | NG | 8.09 | 0.13 | |
| BB3113 | bcaP braB brnQ | 53.5 | 17.7 | 13.6 | |
| BB3283 | bcaPp10/14 | ND | 3.63 | 0.13 | |
| BB2770 | Wild type | ybgE-lacZ | 19.2 | 3.49 | 1.12 |
| BB2870 | liv | NG | 3.25 | 0.87 | |
| BB3114 | bcaP braB brnQ | 22.8 | 6.25 | 9.76 | |
| BB3284 | bcaPp10/14 | ND | 2.24 | 1.00 | |
Cells were grown in TSS glucose-ammonium medium with or without mixtures of ILV (200 μg/ml) and 13 amino acids. The β-galactosidase specific activity was assayed and is expressed in Miller units. All values are averages from at least two experiments, and the mean errors did not exceed 30%. The data for strains BB2505 and BB2770 were taken from previous publications (22, 23). NG, no growth; ND, not determined.
Indeed, the ability of an ILV-containing amino acid mixture to activate CodY, as detected by expression of the negatively regulated bcaP-lacZ fusion, was much reduced by simultaneous inactivation of BcaP, BraB, and BrnQ, despite the presence of high concentrations of ILV in the medium (Table 5, strain BB3113; in these experiments, the bcaP promoter served only as a reporter for CodY activity without any connection to BcaP function as a BCAA permease). A reduction in CodY-mediated repression of the bcaP promoter was also observed in the bcaP braB brnQ strain if only ILV were added to TSS medium (Table 5). Very similar data were obtained for another CodY-regulated fusion, ybgE-lacZ (Table 5). It is likely that the remaining CodY-mediated regulation in triple permease mutants was due to Leu uptake and residual uptake of Ile and Val.
The roles of different ILV permeases in BCAA uptake are likely to depend on the efficiency of their expression. For example, expression of the bcaP gene is virtually abolished in amino acid-rich media due to strong CodY-mediated repression (Table 5) (22). Because expression of the bcaP-lacZ fusion likely correlates with the activity of BcaP, we considered the possibility that this high level of bcaP regulation serves to limit BCAA uptake and, as a result, diminish activation of CodY.
We have introduced simultaneously two mutations (p10 and p14) in the regulatory region of the chromosomal bcaP gene that inactivated the two previously characterized CodY-binding sites within the bcaP promoter (22). Mutation p10 is very similar to the previously described p2 mutation (22) (see Materials and Methods). Both p10/p14 and p2/p14 mutations caused strong derepression of the bcaP promoter even when CodY was highly active (22; data not shown). Our expectation was that the derepressed BcaP would be responsible for more efficient ILV transport and higher activity of CodY. Indeed, using expression of a CodY-regulated fusion as a sensitive indicator, we observed 1.5- to 2-fold stronger repression of the bcaPp+-lacZ and ybgEp+-lacZ fusions in the minimal medium containing ILV (Table 5, strains BB3283 and BB3284). We did not find a similar effect in the medium containing ILV and 13 other amino acids (Table 5). Thus, activities of BraB and BrnQ are apparently sufficient to provide enough ILV uptake to fully activate CodY in the 16 aa-containing medium even if the bcaP gene is repressed; interestingly, brnQ expression is known to be stimulated in amino acid-containing media (31). However, if only ILV are added to the minimal medium, activity of CodY not only determines the efficiency of bcaP repression but is also modulated, in a negative-feedback loop, by the level of bcaP expression.
Role of the azlCD genes in ILV uptake.
We have shown previously that the azlCD genes of the azlBCDEF operon encode a likely exporter of 4-azaleucine (31). Later, the role of Corynebacterium glutamicum genes very similar to azlCD in the export of ILV and methionine was demonstrated experimentally (44, 45). To test whether the activity of the putative AzlCD exporter affects the cells' ability to utilize Ile or Val as the sole nitrogen source or the ability of exogenous ILV to activate CodY, we constructed a deletion that removed parts of both azlC and azlD (see Materials and Methods). Note that azlCD is coregulated with brnQ (azlE) (31).
We could not detect any significant effect of the azlCD deletion on expression of CodY-regulated genes in the presence of ILV in either an azlB+ or an azlB-null background (data not shown). Thus, the azlCD genes are unlikely to contribute significantly to the maintenance of the cellular ILV pool. As a corollary, it means that the phenotypic effects of the brnQ deletion used in this work, which also encompassed the azlCD genes, were unlikely to be affected by the lack of AzlC and AzlD.
Role of BcaP in the transport of threonine and other amino acids.
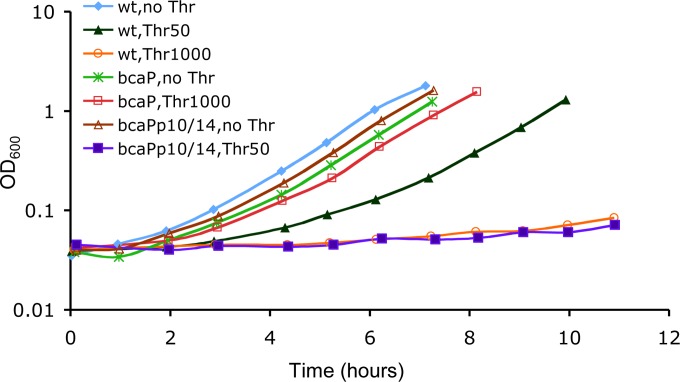
In accord with previous reports (46, 47), threonine, in concentrations of greater than 50 μg/ml, inhibited the growth of wild-type B. subtilis cells (Fig. 4). Cells containing the bcaP-null mutation lost sensitivity to threonine even at concentrations as high as 1 mg/ml; on the other hand, growth inhibition due to threonine addition was magnified by increased bcaP expression caused by the promoter region mutations p10 and p14, which relieve CodY-mediated repression of bcaP (Fig. 4). The results indicate that BcaP may be involved in threonine uptake. As reported (47), the addition of Val (40 μg/ml) reversed the threonine toxicity (data not shown), although neither the mechanism of inhibition nor the mechanism of Val action have been investigated further.
FIG 4.
Growth inhibition by threonine. Cells of strain SMY (wild type), BB2726 (bcaP), or BB3198 (bcaPp10/14) were grown overnight in TSS glucose-ammonium minimal medium and then diluted 100-fold in the same medium with or without threonine (50 or 1,000 μg/ml) as indicated. Each growth experiment was performed at least twice, and the results of a representative experiment are shown.
The bcaP-null mutant cells were as sensitive to inhibitory concentrations of serine (37), as were wild-type cells, and were not defective in the ability to utilize alanine or asparagine as the sole nitrogen source. Thus, although these three amino acids were able to compete with the BcaP-mediated Ile uptake, BcaP, even if it is involved in transport of serine, alanine, or asparagine, is not the only permease for these amino acids.
A bcaP homolog (25 to 28% identity) in B. licheniformis and B. pumilus is located between the gabT and gabD genes that are involved in γ-aminobutyrate (GABA) utilization in B. subtilis (48). This suggests that BcaP may be involved in GABA uptake. Indeed, the known gabP-encoded GABA permease is not the only GABA permease in B. subtilis cells (48, 49). However, in contrast to a gabP-null mutation, a bcaP-null mutation did not affect expression from the gabT promoter, which is dependent on GABA uptake (48); the bcaP mutation also did not exacerbate the gabT expression defect in the gabP mutant strain (unpublished results). Thus, BcaP is unlikely to be involved in GABA uptake in B. subtilis.
DISCUSSION
In this work, we have identified three permeases, BcaP, BraB, and BrnQ, that are essential collectively for the high-affinity uptake of Ile and Val in B. subtilis and are likely involved in the uptake of Leu as well. BcaP contains 465 amino acids and has several paralogs in the B. subtilis genome, including one close relative, YfnA, that is 57% identical to BcaP. The substrate specificity of YfnA is not known. BraB and BrnQ have 445 and 440 amino acids, respectively, and are 49% identical; they have no other paralogs in the B. subtilis genome. All of them are predicted to be integral membrane proteins with multiple membrane-spanning domains.
The BcaP permease is required for utilization of Ile or Val as the sole nitrogen source and is apparently the most efficient Ile/Val permease under these conditions. BcaP appears to be also involved in the uptake of high concentrations of threonine that inhibit growth of B. subtilis cells and may be an amino acid permease of rather broad specificity, because several amino acids, albeit at 100-fold excess, were able to prevent Ile uptake. L. lactis BcaP, a permease that is distantly related to B. subtilis BcaP, was shown to transport methionine in addition to ILV (38); however, B. subtilis BcaP apparently is not capable of using methionine as an efficient substrate because even at 100-fold excess it was only a partial inhibitor of Ile uptake. Both BraB and BrnQ are involved in the uptake of Ile and Val and appear to be specific only for BCAAs, although Leu is apparently not a substrate of BrnQ.
Ile and Val also serve as the substrates for at least one more, low-affinity B. subtilis permease. Moreover, at least one permease, other than BcaP, BraB, or BrnQ, is important for Leu uptake. YvbW, a putative amino acid permease, whose expression is apparently regulated by a Leu-specific T-box mechanism (50), is a prime candidate for such a permease. Multiple BCAA permeases have been identified previously in other bacteria (51).
Simultaneous inactivation of all three BCAA permeases identified in this work strongly reduced B. subtilis CodY activity in ILV-containing media. Thus, the levels of activity and expression of these permeases under different growth conditions determine the efficiency of CodY-mediated transcriptional regulation. Expression of the brnQ gene is subject to efficient negative regulation by the AzlB protein, a member of the Lrp/AsnC family of transcriptional regulators (31). When derepressed, BrnQ can substitute for BcaP in transporting enough Ile or Val to allow them to serve as the sole nitrogen source. Unfortunately, the mechanism by which AzlB activity is regulated remains unknown; though it is increased in the presence of an amino acid mixture in the medium, it is not affected by ILV or methionine only (unpublished results).
The brnQ gene is a part of the azlBCDEF operon but may also be expressed from its own promoter (31). Interestingly, the regulatory regions of both the azlBCDEF operon and the braB gene contain rather strong CodY-binding sites (2). However, expression of neither azlB-lacZ (2) nor braB-lacZ nor brnQ-lacZ fusions (data not shown) was affected by a null mutation in the codY gene under the growth conditions tested.
The bcaP (yhdG) gene is one of the genes most highly repressed by CodY (1, 3, 22). Poor expression of BcaP in amino acid-rich media may explain why ILV are utilized only in stationary phase, i.e., when CodY activity diminishes due to exhaustion of other amino acids and BcaP-mediated transport of ILV apparently resumes (52). Importantly, such a pattern of ILV utilization should extend the time when CodY is maintained in at least a partially active state. More generally, the negative effect of CodY on the expression of BcaP, a permease that contributes to supplying cells with ILV, constitutes an indirect negative autoregulatory loop that allows fine-tuning of ILV-responsive CodY activity.
ACKNOWLEDGMENTS
I am grateful to A. L. Sonenshein for encouragement, helpful discussions, and careful reading of the manuscript.
This study was supported by a research grant (GM042219) from the U.S. National Institute of General Medical Sciences to A. L. Sonenshein.
The content is solely the responsibility of the author and does not represent the official view of the National Institutes of Health or the NIGMS.
REFERENCES
- 1.Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol 185:1911–1922. doi: 10.1128/JB.185.6.1911-1922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky BR, Sonenshein AL. 2013. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc Natl Acad Sci U S A 110:7026–7031. doi: 10.1073/pnas.1300428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinsmade SR, Alexander EL, Livny J, Stettner AI, Segre D, Rhee KY, Sonenshein AL. 2014. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc Natl Acad Sci U S A 111:8227–8232. doi: 10.1073/pnas.1321308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slack FJ, Serror P, Joyce E, Sonenshein AL. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol 15:689–702. [DOI] [PubMed] [Google Scholar]
- 5.Fisher SH. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol Microbiol 32:223–232. [DOI] [PubMed] [Google Scholar]
- 6.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol 8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol 5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 8.Geiger T, Wolz C. 2014. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int J Med Microbiol 304:150–155. doi: 10.1016/j.ijmm.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Dahlsten E, Korkeala H, Lindstrom M. 2014. Positive regulation of botulinum neurotoxin gene expression by CodY in Clostridium botulinum ATCC 3502. Appl Environ Microbiol 80:7651–7658. doi: 10.1128/AEM.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roux A, Todd DA, Velazquez JV, Cech NB, Sonenshein AL. 2014. CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J Bacteriol 196:1184–1196. doi: 10.1128/JB.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serror P, Sonenshein AL. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol Microbiol 20:843–852. doi: 10.1111/j.1365-2958.1996.tb02522.x. [DOI] [PubMed] [Google Scholar]
- 12.Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J Biol Chem 281:11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]
- 13.Guedon E, Serror P, Ehrlich SD, Renault P, Delorme C. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol Microbiol 40:1227–1239. doi: 10.1046/j.1365-2958.2001.02470.x. [DOI] [PubMed] [Google Scholar]
- 14.Petranovic D, Guedon E, Sperandio B, Delorme C, Ehrlich D, Renault P. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol Microbiol 53:613–621. doi: 10.1111/j.1365-2958.2004.04136.x. [DOI] [PubMed] [Google Scholar]
- 15.Shivers RP, Sonenshein AL. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol 53:599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 16.den Hengst CD, Curley P, Larsen R, Buist G, Nauta A, van Sinderen D, Kuipers OP, Kok J. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J Bacteriol 187:512–521. doi: 10.1128/JB.187.2.512-521.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levdikov VM, Blagova E, Colledge VL, Lebedev AA, Williamson DC, Sonenshein AL, Wilkinson AJ. 2009. Structural rearrangement accompanying ligand binding in the GAF domain of CodY from Bacillus subtilis. J Mol Biol 390:1007–1018. doi: 10.1016/j.jmb.2009.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handke LD, Shivers RP, Sonenshein AL. 2008. Interaction of Bacillus subtilis CodY with GTP. J Bacteriol 190:798–806. doi: 10.1128/JB.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brinsmade SR, Sonenshein AL. 2011. Dissecting complex metabolic integration provides direct genetic evidence for CodY activation by guanine nucleotides. J Bacteriol 193:5637–5648. doi: 10.1128/JB.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villapakkam AC, Handke LD, Belitsky BR, Levdikov VM, Wilkinson AJ, Sonenshein AL. 2009. Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids. J Bacteriol 191:6865–6876. doi: 10.1128/JB.00818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belitsky BR, Sonenshein AL. 2011. Contributions of multiple binding sites and effector-independent binding to CodY-mediated regulation in Bacillus subtilis. J Bacteriol 193:473–484. doi: 10.1128/JB.01151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belitsky BR, Sonenshein AL. 2011. Roadblock repression of transcription by Bacillus subtilis CodY. J Mol Biol 411:729–743. doi: 10.1016/j.jmb.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atkinson MR, Wray LV Jr, Fisher SH. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J Bacteriol 172:4758–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slack FJ, Mueller JP, Strauch MA, Mathiopoulos C, Sonenshein AL. 1991. Transcriptional regulation of a Bacillus subtilis dipeptide transport operon. Mol Microbiol 5:1915–1925. doi: 10.1111/j.1365-2958.1991.tb00815.x. [DOI] [PubMed] [Google Scholar]
- 26.Fisher SH, Rohrer K, Ferson AE. 1996. Role of CodY in regulation of the Bacillus subtilis hut operon. J Bacteriol 178:3779–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belitsky BR, Sonenshein AL. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J Bacteriol 190:1224–1236. doi: 10.1128/JB.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 30.Belitsky BR, Sonenshein AL. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol 180:6298–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belitsky BR, Gustafsson MC, Sonenshein AL, Von Wachenfeldt C. 1997. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J Bacteriol 179:5448–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller JP, Bukusoglu G, Sonenshein AL. 1992. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J Bacteriol 174:4361–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeDeaux JR, Grossman AD. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol 177:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belitsky BR. 2011. Indirect repression by Bacillus subtilis CodY via displacement of the activator of the proline utilization operon. J Mol Biol 413:321–336. doi: 10.1016/j.jmb.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel RA, Haiech J, Denizot F, Errington J. 1997. Isolation and characterization of the lacA gene encoding beta-galactosidase in Bacillus subtilis and a regulator gene, lacR. J Bacteriol 179:5636–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang TZ, Moszer I, Medigue C, Danchin A. 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155:1758–1775. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandeyar MA, Zahler SA. 1986. Chromosomal insertions of Tn917 in Bacillus subtilis. J Bacteriol 167:530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Hengst CD, Groeneveld M, Kuipers OP, Kok J. 2006. Identification and functional characterization of the Lactococcus lactis CodY-regulated branched-chain amino acid permease BcaP (CtrA). J Bacteriol 188:3280–3289. doi: 10.1128/JB.188.9.3280-3289.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.den Hengst CD, van Hijum SA, Geurts JM, Nauta A, Kok J, Kuipers OP. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J Biol Chem 280:34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- 40.Fink PS. 1993. Biosynthesis of the branched-chain amino acids, p 307–317. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and other Gram-positive bacteria. American Society for Microbiology, Washington, DC. [Google Scholar]
- 41.Obermeier N, Poralla K. 1976. Some physiological functions of the l-leucine dehydrogenase in Bacillus subtilis. Arch Microbiol 109:59–63. doi: 10.1007/BF00425113. [DOI] [PubMed] [Google Scholar]
- 42.Debarbouille M, Gardan R, Arnaud M, Rapoport G. 1999. Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol 181:2059–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL. 2010. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J Bacteriol 192:6357–6368. doi: 10.1128/JB.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennerknecht N, Sahm H, Yen MR, Patek M, Saier MH Jr, Eggeling L. 2002. Export of l-isoleucine from Corynebacterium glutamicum: a two-gene-encoded member of a new translocator family. J Bacteriol 184:3947–3956. doi: 10.1128/JB.184.14.3947-3956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trotschel C, Deutenberg D, Bathe B, Burkovski A, Kramer R. 2005. Characterization of methionine export in Corynebacterium glutamicum. J Bacteriol 187:3786–3794. doi: 10.1128/JB.187.11.3786-3794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamb DH, Bott KF. 1979. Threonine inhibition of growth of Bacillus subtilis: positive selection for isoleucine auxotrophy. J Gen Microbiol 111:433–435. doi: 10.1099/00221287-111-2-433. [DOI] [PubMed] [Google Scholar]
- 47.Lamb DH, Bott KF. 1979. Inhibition of Bacillus subtilis growth and sporulation by threonine. J Bacteriol 137:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belitsky BR, Sonenshein AL. 2002. GabR, a member of a novel protein family, regulates utilization of γ-aminobutyrate in Bacillus subtilis. Mol Microbiol 45:569–583. doi: 10.1046/j.1365-2958.2002.03036.x. [DOI] [PubMed] [Google Scholar]
- 49.Ferson AE, Wray LV Jr, Fisher SH. 1996. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol Microbiol 22:693–701. doi: 10.1046/j.1365-2958.1996.d01-1720.x. [DOI] [PubMed] [Google Scholar]
- 50.Wels M, Groot Kormelink T, Kleerebezem M, Siezen RJ, Francke C. 2008. An in silico analysis of T-box regulated genes and T-box evolution in prokaryotes, with emphasis on prediction of substrate specificity of transporters. BMC Genomics 9:330. doi: 10.1186/1471-2164-9-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoshino T, Kose-Terai K, Uratani Y. 1991. Isolation of the braZ gene encoding the carrier for a novel branched-chain amino acid transport system in Pseudomonas aeruginosa PAO. J Bacteriol 173:1855–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liebs P, Riedel K, Graba JP, Schrapel D, Tischler U. 1988. Formation of some extracellular enzymes during the exponential growth of Bacillus subtilis. Folia Microbiol (Praha) 33:88–95. doi: 10.1007/BF02928073. [DOI] [PubMed] [Google Scholar]
- 53.Dedonder RA, Lepesant JA, Lepesant-Kejzlarova J, Billault A, Steinmetz M, Kunst F. 1977. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl Environ Microbiol 33:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]