Figure 1.
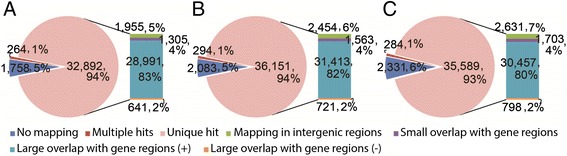
Mapping of de novo assembled transcripts of the three Chinese wild Vitis, BH (A), HN (B), and S (C), to the reference PN40024 genome. No mapping: Contigs not mapped; Multiple hits: Contigs mapped to multiple genomic locations; Unique hit: Contigs mapped to unique genomic locations; Mapping in intergenic regions: Contigs mapped to intergenic regions; Small overlap with gene regions: Contigs mapped to gene regions with low overlapping (<90% of contig length); Large overlap with gene regions (+): Contigs mapped to gene regions in sense directions with high overlapping (≥90% of contig length) ; Large overlap with gene regions (−): Contigs mapped to gene regions in antisense directions with high overlapping (≥90% of contig length). BH, V. pseudoreticulata accession “Baihe-13-1”; HN, V. pseudoreticulata accession “Hunan-1”; S, V. quinquangularis accession “Shang-24”.
