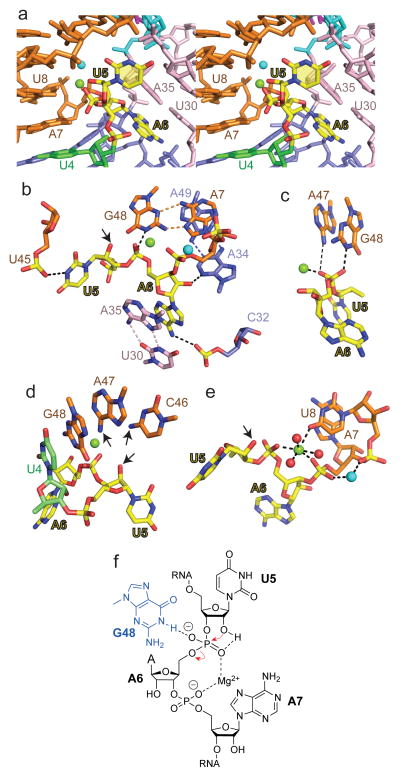
Figure 4. Intermolecular contacts and bound metal ions at the U5-A6 cleavage site step in the structure of the dU5-containing env22 twister ribozyme and chemical drawing showing the structure-derived, chemically important entities for the env22 ribozyme self-cleavage mechanism.
a, A stereo view of the U5-A6 step (in yellow) nestled within the major groove of stem P2 (in orange). Bound Mg2+ and an unassigned cation within the U5-A6-A7 segment are shown in green and cyan. b, A view of intermolecular hydrogen bonds involving the U5-A6 step. Also shown is the relative positioning of A7•G48, U30•A35 and A34•A39 non-canonical pairs. c, View showing hydrogen bonding by A47 (weak) and G48 (strong) and coordination by bound Mg2+ cation to the non-bridging oxygens of the phosphate at the U5-A6 step. d, View showing the positioning of G48 directed towards the non-bridging oxygens of the phosphate at the U5-A6 step, and the positioning of C46 and A47 directed towards the modeled 2′-O of U5. Arrows indicate amino groups of C46 and A47 that are directed towards the 2′-O of U5. e, Bound cations at the U5-A6-A7 step. The bound Mg2+ and unassigned cation are in green and cyan respectively, while water molecules are in red. f, Schematic highlighting in-line attack of the U5 2′-OH, Mg2+ coordination to U5-A6 and A6-A7 phosphodiester groups.