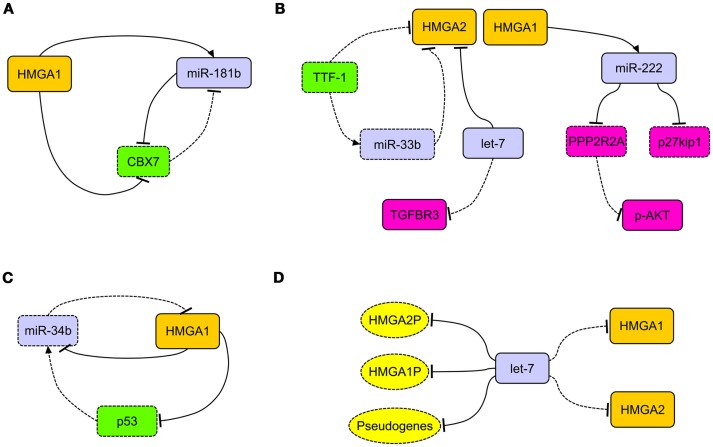
Figure 1.
The network of HMGA regulatory mechanisms in human carcinomas. (A) In breast carcinoma, HMGA1 is able to simultaneously repress the expression of CBX7 and induce the expression of miR-181b. This latter, together with CBX7, takes part to a reciprocal regulation. (B) HMGA2, overexpressed in lung carcinomas, acts as competing endogenous RNA for let-7, allowing the activation of the TGF-beta signaling through the upregulation of TGFBR3. Decreased expression of TTF-1 in lung carcinomas allows the overexpression of HMGA2 protein directly, by releasing the transcriptional block on its promoter, and indirectly, by removing the translational block due to miR-33b. HMGA1 is able to induce the expression of miR-222, which in turn can target p27kip1 and PPP2R2A, then activating the AKT signaling. (C) The loss of miR-34b expression in cancer cells allows the overexpression of HMGA1, which in turn alters the miR-34b pathway by repressing it and its inducer p53. (D) The presence of several let-7 binding sites in the 3′ untranslated regions of HMGA1, HMGA2, and relative pseudogenes (HMGA1P, HMGA2P) alters the epigenetic modulation of HMGA2 and HMGA1 themselves (respectively), allowing their overexpression after decoy of let-7 microRNA. Dashed rectangles/ovals and lines represent decreased expression or loss of regulatory action, respectively.