Figure 1. CD22 Trans-splicing with an RNA Trans-splicing Molecule (RTM).
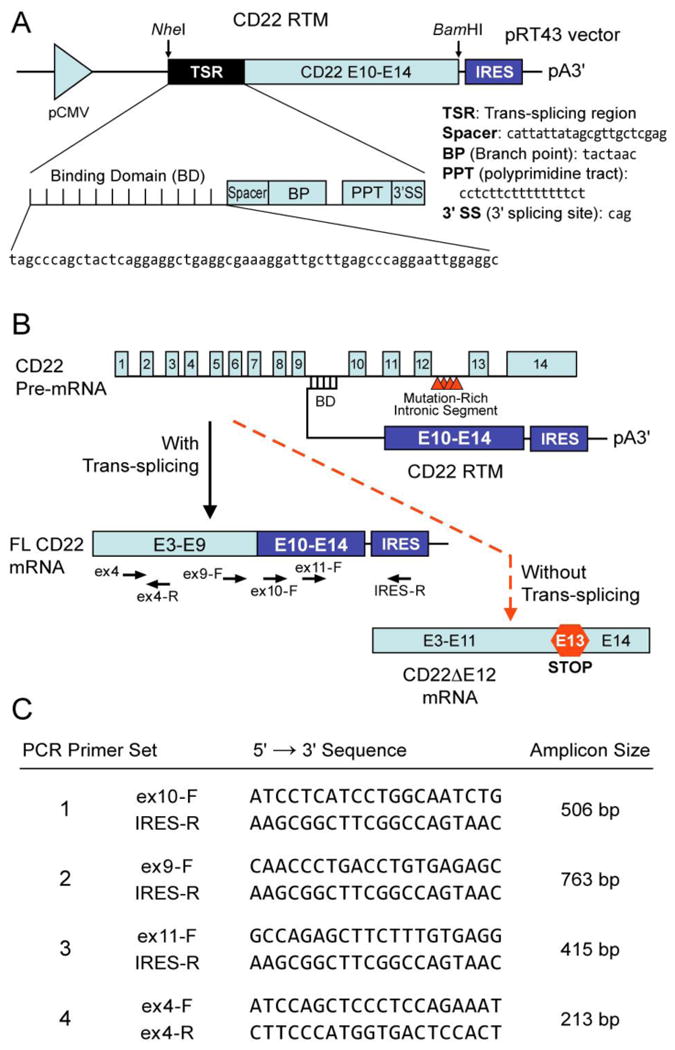
[A] RTMs have 3 main elements, 1) an anti-sense binding domain (BD) within a trans-splicing region (TSR) which serves as a “targeting moiety” that confers specificity by tethering the RTM to its target pre-mRNA; 2) a 3′ and/or 5′ splice site; and 3) a coding sequence to be trans-spliced, which can re-write most of the targeted pre-mRNA by replacing one or numerous exons anywhere in a message. Depicted is a schematic representation of the CD22 RTM that contains a BD targeting sequence complementary to a 60 nucleotide region close to the 5′ splice site of CD22 Intron 9 of the human CD22 gene, a 3′ Splice Site (3′SS) and the coding regions of the human CD22 exons 10-14. [B] Spliceosome-mediated RNA trans-splicing to replace the deleted CD22 exon 12 (E12) using a CD22 RTM. This RTM was designed to bind to the CD22 pre-mRNA via conventional Watson-Crick base pairing and replace via trans-splicing the downstream mutation-rich segment of intron 12 and remaining segments in leukemia cells with CD22ΔE12 with wildtype CD22 exons 10-14 and thereby prevent the generation of the Cis-spliced aberrant CD22ΔE12 product. The trans-splicing RTM vector was created by cloning CD22 exons 10-14 as the coding sequence to be trans-spliced and the elements of the 3′ trans-splicing arm into a pIRES2-AcGFP1 vector (Clontech) followed by removal the IRES/GFP sequences from the vector. When trans-splicing occurs, a corrected full-length CD22 mRNA is created from the natural CD22ΔE12 gene, which contains intronic mutations that are associated with Exon 12 deletion. The corrected mRNA would be expressed under the endogenous regulation of the cell.
