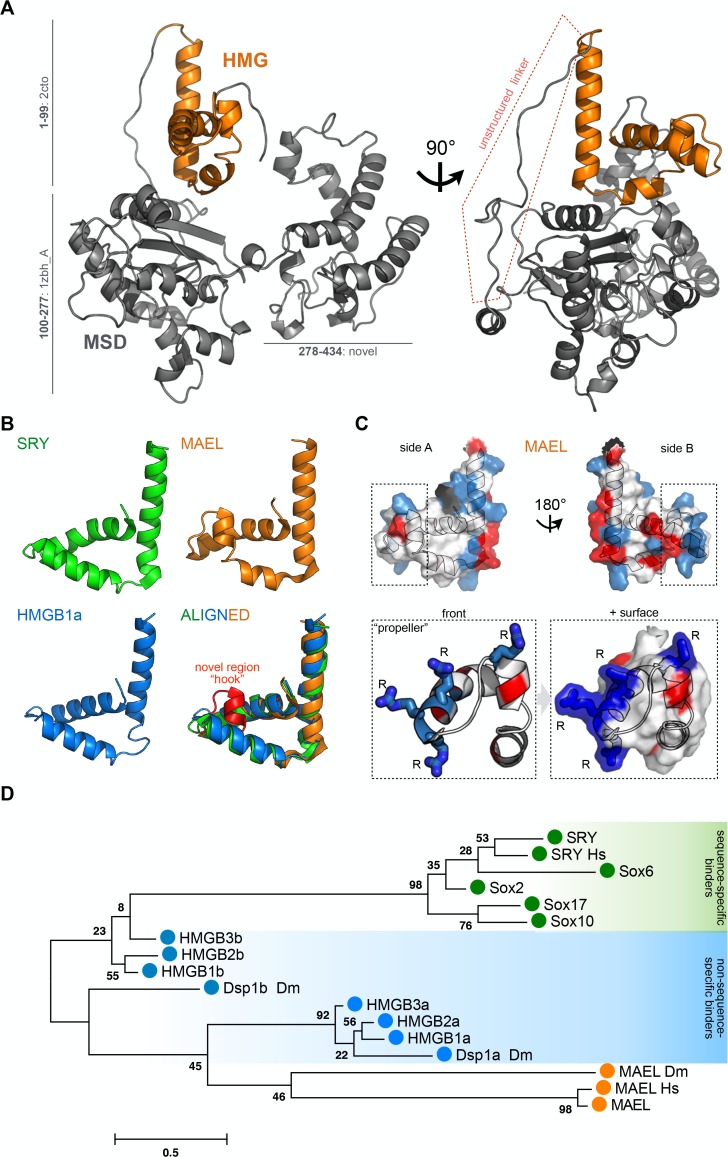
Fig 1. MAEL HMG-box domain structural and sequence considerations.
(A) Tertiary structure prediction of mouse MAEL protein. MAEL HMG-box domain (orange) is positioned on top of the Maelstrom Specific Domain (MSD, grey) linked by unstructured linker (highlighted in red). The model was generated with Robetta software by either de novo modeling or with established structures as templates. Residue ranges and parent PDB ID numbers are shown. (B) Comparison of MAEL HMG-box and canonical HMG-box domains. Determined structures of candidate HMG-box proteins (SRY: 1j46—sequence specific binding; HMGB1a: 1ckt—structure specific binding; MAEL: 2cto—unknown binding) were visualized and structures aligned in PyMOL. MAEL HMG-box domain has a conserved canonical L-shape fold but with a bend in helix-2, creating a novel region termed “hook” (red). (C) Distribution of charged residues of mouse MAEL HMG-box domain. Positive residues—Arg, Lys, His are blue; negative residues—Asp, Glu are red. Charged residues are concentrated on side B. Unlike other HMG-box domains, in MAEL three arginine (R) residues are concentrated at the end of the helix-1, and protrude outwards, forming a “propeller”-like shape. (D) Phylogenic comparison of MAEL HMG-box domain with well-studied candidate HMG-box domains from sequence-specific (single HMG-box, SRY, Sox) and non-sequence-specific (two HMG-boxes, HMGB’s, Dsp1) groups. Mouse sequences were used unless otherwise noted (Dm: Drosophila melanogaster, Hs: Homo sapiens). The phylogenetic tree was generated using maximum likelihood method in MEGA6 software. Values next to the branches describe percentage of trees where associated sequences group together (n = 1000). The branch length scale is in substitutions per site. MAEL HMG-box domain forms a new branch most closely related to the domain A of non-sequence specific HMG-box proteins.