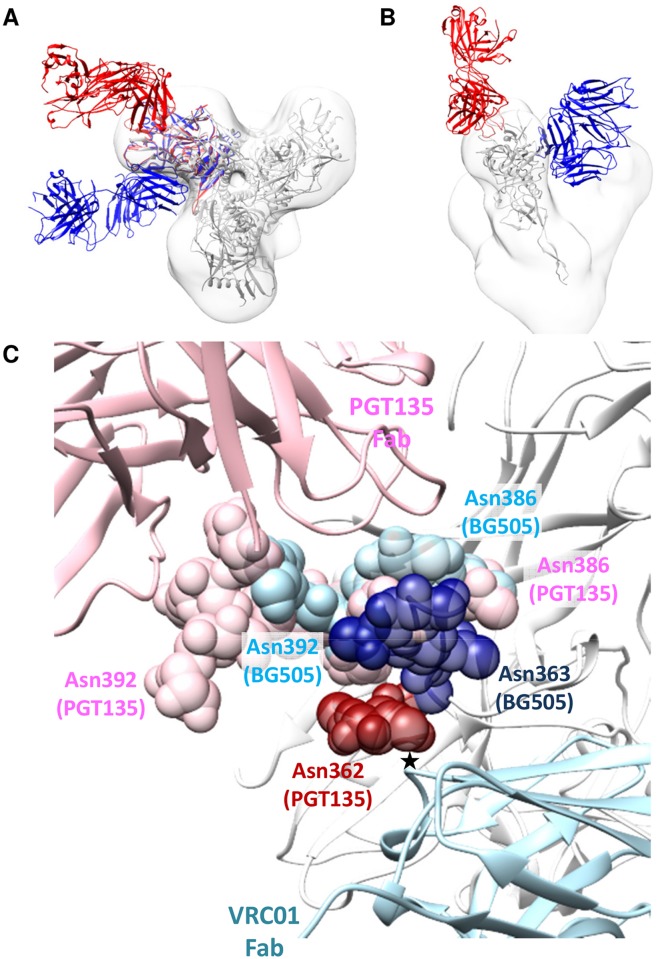
Fig 6. Comparison of PGT135 and VRC01 binding to HIV-1 gp120.
(A) View down the trimer three-fold axis showing a superimposition of PGT135 Fab in complex with the clade B JRFL gp120 core (PDB ID: 4jm2; red), and of VRC01 Fab in complex with the clade A/E 93TH057 gp120 core (PDB ID: 3ngb; blue), each aligned onto the crystal structure of the BG505 SOSIP.664 trimer (PDB ID: 4tvp; white). In addition, the same BG505 SOSIP.664 crystal structure is fit into the negative-stain EM reconstruction of JR-FL Env trimer in complex with PGT151 (EMD-5919; white). The resulting model clearly shows that PGT135 and VRC01 do not sterically block binding of each other. (B) Side view of (A) with only one gp120 monomer displayed for clarity. (C) Detailed view of the key glycans (spheres) that were resolved in the PGT135-gp120 structure, and of the same glycans from the BG505 SOSIP.664 crystal structure. The presumed steric clash between glycan on Asn362 (and perhaps also Asn363) and the CDR H2 loop of VRC01 is marked with an asterisk. The gp120 subunit of the BG505 SOSIP.664 trimer is displayed as white ribbons. The glycans depicted were limited to the components resolved in the crystal structures (GlncNAc1 for Asn362, GlncNAc2Man1 for Asn386 and GlncNAc2Man6 for Asn392 in the PGT135-gp120 structure; GlncNAc2 for Asn362, GlncNAc2 for Asn386 and GlncNAc2 for Asn392 in the BG505 SOSIP.664 trimer structure), and hence their sizes are underestimated compared to trimers produced in 293F cells or present on viruses.