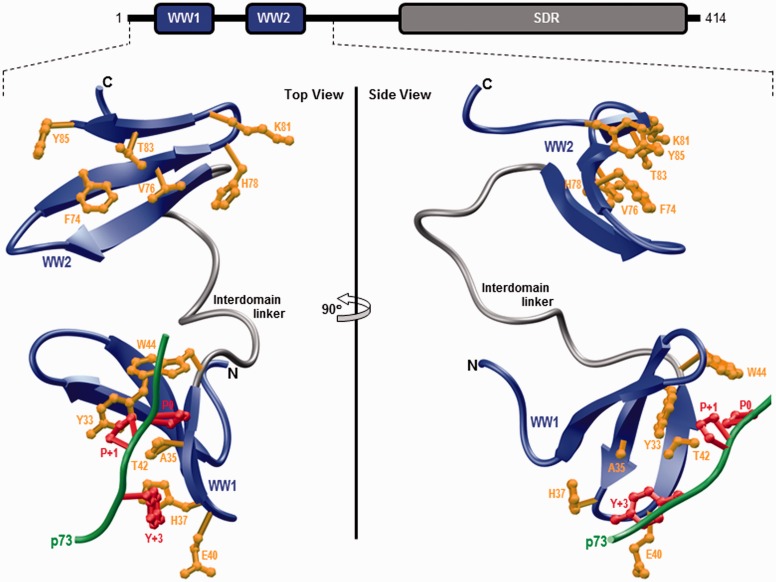
Figure 1.
Ribbon representation of the structural model of WW1–WW2 tandem module of WWOX in complex with the p73 peptide containing the PPXY motif bound to the WW1 domain. Two alternative orientations related by a 90° rotation about the vertical axis are depicted for the inquisitive eye. In each case, the WW domains are shown in blue with the interdomain linker depicted in gray and the bound peptide is colored green. The side chain moieties of residues (yellow) within the WW1 domain engaged in intermolecular contacts with the consensus residues (red) within the PPXY motif of p73 peptide are shown. For comparison, the side chain moieties of structurally equivalent residues (yellow) within the putative ligand binding groove of WW2 domain are also depicted. Note that the modular architecture of WWOX is overlaid to indicate the relative locations of the N-terminal WW1–WW2 tandem module and the C-terminal SDR domain. The structural model of the WW1–WW2 tandem module was built using the MODELLER software based on homology modeling.43 The unliganded model was obtained using the NMR structure of WW1–WW2 tandem module of FBP21 pre-mRNA splicing factor as a template (PDBID 2JXW). To obtain the liganded model, the 12-mer p73 peptide (HCTPPPPYHADP) was docked onto the WW1 domain within the unliganded structure of WW1–WW2 tandem module in a 1:1 stoichiometry using the NMR structure of the homologous WW domain of YAP bound to a peptide containing the PPXY motif as a template (PDBID 1JMQ). The structural model was rendered using RIBBONS.44 (A color version of this figure is available in the online journal.)