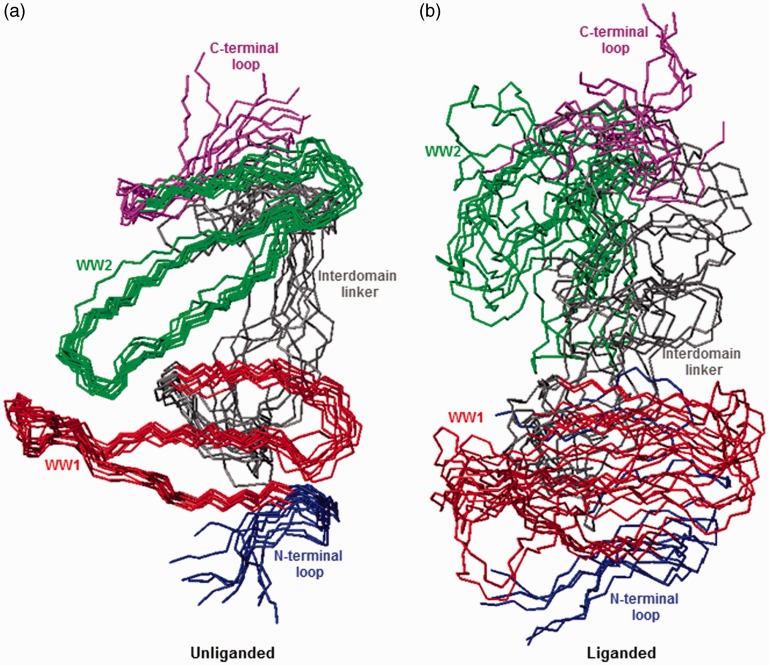
Figure 2.
Superimposition of simulated structures as derived from MD analysis conducted on the structural model of WW1–WW2 tandem module of WWOX alone (unliganded) and in complex with p73 peptide containing the PPXY motif bound to the WW1 domain (liganded). Note that the superimposed structures for the unliganded (a) and liganded (b) WW1–WW2 tandem module were obtained at 200-ns time intervals over a simulation time of 2 µs. All 10 structures were superimposed with respect to the backbone atoms (N, Cα, and C) of the core regions of WW1 (residues 22–43) and WW2 (residues 63–84) domains. In each case, the constituent WW1 (residues 22–43) and WW2 (residues 63–84) domains are, respectively, colored red and green, while the N-terminal loop (residues 16–21), interdomain linker (residues 44–62), and C-terminal loop (residues 85–91) are, respectively, shown in blue, gray, and magenta. In (b), the p73 peptide is not shown for clarity. All MD simulations were performed with the GROMACS software48 using the integrated AMBER99SB-ILDN force field.49,50 Structural snapshots of WW1–WW2 tandem module taken at various time intervals during the course of MD simulations were superimposed using MOLMOL.51 (A color version of this figure is available in the online journal.)