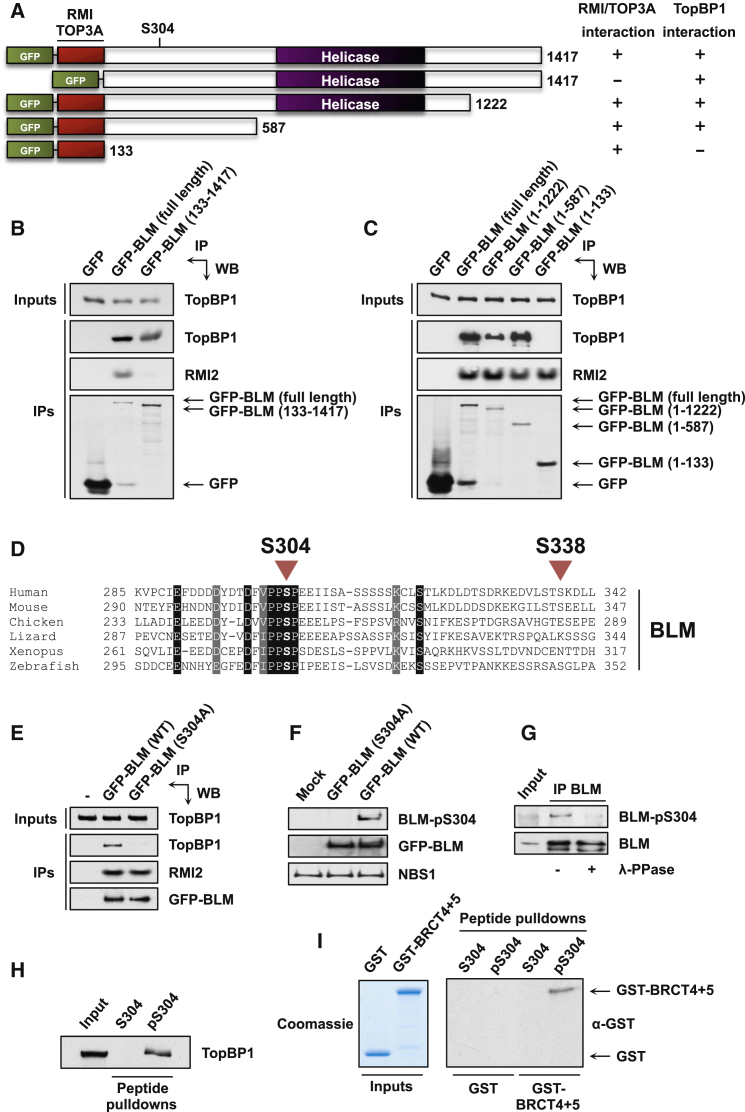
Figure 2.
BLM Ser304 Phosphorylation Mediates Direct Binding to TopBP1-BRCT5
(A) Schematic of the GFP-tagged BLM constructs used in this study.
(B) The N-terminal 132 residues of BLM are required for binding to TOP3A and RMI2 but not TopBP1. Pull-downs were carried out from 293FT cells transiently transfected with the indicated plasmids.
(C) The binding site for TopBP1 is located within residues 133–587 of BLM. RMI2 is a positive control for binding to the N terminus of BLM.
(D) Sequence alignment showing the evolutionary conservation of the BLM region containing Ser304 and Ser338.
(E) Mutation of Ser304 specifically abrogates binding to TopBP1. Pull-downs were carried out from U2OS cells stably expressing the indicated proteins.
(F) The BLM-pS304 antibody does not recognize BLM-S304A. 293FT cells were transiently transfected with the indicated plasmids. NBS1 is a loading control.
(G) Ser304 is phosphorylated in vivo. BLM immunoprecipitates from U2OS cells were mock treated or treated with lambda phosphatase (λ-PPase).
(H) BLM residues 297–311 are sufficient for interaction with TopBP1 when Ser304 is phosphorylated. Streptavidin beads were incubated with biotinylated peptides before addition to HeLa nuclear extracts for pull-downs.
(I) TopBP1 BRCT domains 4 and 5 interact directly with BLM peptides phosphorylated on Ser304. Streptavidin beads were incubated with biotinylated peptides before mixing with GST-tagged BRCT domains 4 and 5 or GST alone. See also Figure S2.