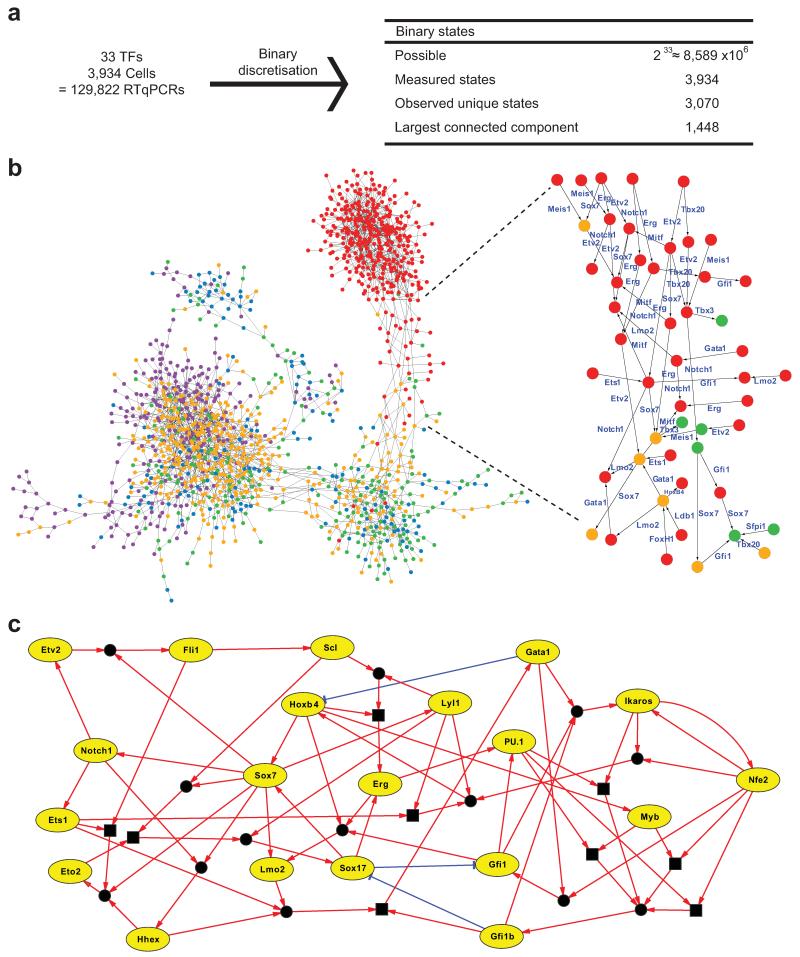
Figure 3. Regulatory network synthesis from single-cell expression profiles.
(a) Discretisation of 3,934 expression profiles for 33 TFs yields 3,070 unique binary states, 1448 of which can be connected by single-gene changes to yield a state graph. (b) Representation of resulting state graph, coloured by first embryonic stage appearing in each state. Blue, PS; green, NP; orange, HF; red, 4SG; purple, 4SFG−. Magnification of fate transition towards 4SG states, with for example Sox7 expression switching off along all routes. (c) Representation of synthesised asynchronous Boolean network models for core network of 20 TFs. Green edges indicate activation; red edges indicate repression. Square boxes represent AND operations. Circles connecting edges indicate multiple update rules.